Abstract
Misfit sidechains in protein crystal structures are a stumbling block in using those structures to direct further scientific inference. Problems due to surface disorder and poor electron density are very difficult to address, but a large class of systematic errors are quite common even in well-ordered regions, resulting in sidechains fit backwards into local density in predictable ways. The MolProbity web site is effective at diagnosing such errors, and can perform reliable automated correction of a few special cases such as 180° flips of Asn or Gln sidechain amides, using all-atom contacts and H-bond networks. However, most at-risk residues involve tetrahedral geometry, and their valid correction requires rigorous evaluation of sidechain movement and sometimes backbone shift. The current work extends the benefits of robust automated correction to more sidechain types. The Autofix method identifies candidate systematic, flipped-over errors in Leu, Thr, Val, and Arg using MolProbity quality statistics, proposes a corrected position using real-space refinement with rotamer selection in Coot, and accepts or rejects the correction based on improvement in MolProbity criteria and on χ angle change. Criteria are chosen conservatively, after examining many individual results, to ensure valid correction. To test this method, Autofix was run and analyzed for 945 representative PDB files and on the 50S ribosomal subunit of file 1YHQ. Over 40% of Leu, Val, and Thr outliers and 15% of Arg outliers were successfully corrected, resulting in a total of 3,679 corrected sidechains, or 4 per structure on average. Summary Sentences: A common class of misfit sidechains in protein crystal structures is due to systematic errors that place the sidechain backwards into the local electron density. A fully automated method called “Autofix” identifies such errors for Leu, Val, Thr, and Arg and corrects over one third of them, using MolProbity validation criteria and Coot real-space refinement of rotamers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
One of the challenges in working with crystal structures of proteins is identifying and addressing misfit sidechains. As electron density becomes less defined at lower resolution, some incorrect or impossible sidechain conformers score as favorably as their correct counterparts in traditional model-building and refinement methods, allowing these sidechains to be trapped in the wrong local minimum. Errors in deposited structures may hurt inferences about active site function, misdirect mutagenesis studies, prevent accurate homology modeling, or propagate error in structural bioinformatic studies such as rotamer libraries or pairing preferences of amino acid residues at protein-protein interfaces. If such errors can be easily identified and corrected prior to coordinate deposition, their future negative effects can be avoided.
Though sidechains may be misfit in any number of incorrect conformations, a notable percentage are systematic errors where the end of the sidechain is fit flipped backwards, about 180° from its correct conformation, within electron density with an elongated cross-section but not a clear shape. Leu in particular is often misfit in this manner, seen in database surveys as the decoy rotamers tt* and mp* instead of the correct tp or mt rotamers [1]. The guanidinium group of Arg, as well as the branched-Cβ residues Val/Thr/Ile, exhibit similar patterns of systematic backward misfits. These conformations are reasonable fits to the local electron density, but are not rotameric and are accompanied by clashes with neighboring residues, eclipsed χ angles, distorted geometry, and/or unmet H-bond opportunities. Some of these systematic errors, such as swapped locations of sidechain amide N and O atoms in Asn or Gln residues, can already be identified and fixed automatically through addition of hydrogen atoms and examination of all-atom steric clashes and H-bond networks [2, 3]. However, most misfittings require more extensive movement of the entire sidechain and subsequent re-refinement. As a result, when these errors are addressed at all, it is either by tedious manual correction or by model-building methods still subject to the original sources of error. Successful manual correction of nearly all misfit sidechains for about 30 structures was shown by Arendall et al. [4], with concomitant decreases in steric clashes, Ramachandran outliers, and R and Rfree values.
The breadth and accuracy of structure in the Protein Data Bank [5] (4,025 structures at ≤1.5 Å as of 10/14/08) has yielded much insight into real sidechain conformations observed in protein structures. Programs such as What If [6], OOPS [7], ProCheck [8], and MolProbity [3] use this empirical data to provide validation statistics and identification of local problems in a protein structure. The facilities of these validation suites can be used both to target problematic residues and also to evaluate a proposed correction, either in a user-interactive method or in full automation.
Many software systems include features to avoid or correct fitting errors. Interactive model-building software such as O [7] and Coot [9] provide users with a variety of evaluation and rebuilding tools, which have developed increasingly sophisticated semi-automated aids in recent years but are not meant to be fully automatic. Fragment based loop-building programs such as Xpleo [10] and Loopy [11] are highly effective but typically do not explicitly filter misfit sidechains or backbone from their fragment libraries and so can propagate mistakes. Automated chain-tracing programs like Arp/Warp [12] and Resolve [13] build fewer errors than found in many earlier structures, because they make extensive use of empirical knowledge such as rotamer and Ramachandran distributions. But those errors they do miss remain mostly uncorrected. In our own earlier efforts to extend automated sidechain correction, we developed algorithms that worked well for typical cases, but never succeeded in tuning them to avoid making occasional bad miscorrections (false positives) more often than we were willing to tolerate. Since then, we have therefore concentrated on how to develop a conservative system that can reliably determine when not to make a change.
The current work presents the Autofix method to automatically identify and correct a large fraction of misfit Leu, Val, Thr, and Arg sidechain outliers in crystal structures, with very few false positives. It builds upon many of the available tools, but decides on acceptance by a stringent system of independent validation criteria. It was developed and tested by runs on a set of 945 representative PDB files ranging in resolution from 0.98 to 4.5 Å, and also on the 1YHQ 50S ribosomal subunit. In all, 3,649 sidechain corrections were accepted, a sampling of which were examined to select and validate details of the method.
Materials and methods
Dataset
1,028 representative PDB files were chosen at random from a set of structure-factor files provided by the PHENIX project [14], but required to contain protein and at least one residue of Leu, Val, Thr, or Arg. The associated v2.3 format PDB files were run through MolProbity to add and optimize hydrogens and to correct Asn/Gln/His flips. These modified PDB files were then run through phenix.refine with no refinement, to generate electron density maps for use in the Autofix protocol. 945 of the 1,028 files were used in this study; the remainder were rejected, usually because they failed map generation due to incomplete or oddly formatted Rfree data. Data files varied in resolution from 0.98 to 4.5 Å, with an average of 2.1 Å. In the set of 945 files, 364 structures are <2.0 Å, 348 are between 2.0 and 2.5 Å, 177 are between 2.5 and 3.0 Å, and 56 are ≥3.0 Å resolution. The 945 PDB codes and resolutions are listed in the Supplementary Material.
As a companion study, we also ran the Autofix method on the 50S ribosomal subunit from Haloarcula marismortui (PDBid: 1YHQ) [15]. 1YHQ has a resolution of 2.4 Å, and contains 229 Leu, 313 Val, 206 Thr, and 339 Arg residues. The large number of relevant residues provides an excellent self-consistent test case for comparison with the results from the large dataset analysis. The increased fraction of Arg residues in this structure reflects their major importance in protein/RNA interactions [16].
Electron density map generation
The mtz-format structure-factor files used in this study were generated using PHENIX 1.3-rc2 by running phenix.cif_as_mtz on cif-format structure-factor files available from the PDB. CNS-format electron density maps were generated using PHENIX 1.3-rc2 by running phenix.refine with zero macro cycles of refinement using the published PDB coordinate files and the generated mtz files, resulting in 2Fo-Fc maps that are compatible with Coot.
Autofix methodology overview
-
1.
Identify candidate misfit residues using MolProbity
-
2.
Attempt correction in Coot
-
3.
Rerun MolProbity analysis on proposed correction
-
4.
Accept/reject correction
-
5.
Output improved PDB coordinate file
Outlier candidate identification
Outliers are identified through analysis of MolProbity validation scores for the residue in question. Rotamer scores (rotamericity) and Ramachandran scores are based on smoothed empirical distributions of high-quality, B-factor filtered data in the relevant high-dimensional dihedral-angle space. The reported score is the percentage of reference, high-quality data points that score worse than the residue being evaluated [1, 17]. Hydrogens are added and optimized in Reduce [2] and all-atom contacts calculated with Probe [18]. A simplified per-residue clash overlap score is defined as the largest atomic overlap ≥0.4 Å involving any atom in that residue, or otherwise 0. Cβ deviation is the distance between the deposited Cβ position and an ideal Cβ calculated from the residue’s backbone atoms [17]; it gives a combined measure of angle distortions around the Cα.
Candidates are separated into two categories, defined here as outlier and borderline. The runs of Autofix analyzed both classes of candidates for test purposes, but most analyses reported here, and the methodology currently adopted, use only outliers (see Discussion). Outlier candidates have a rotamericity <1.0% or a near-eclipsed χ angle ± 20°. Borderline candidates have either rotamer score ≤6.0% or a near-eclipsed χ angle ±40°. All initial scores are stored for each candidate residue, for comparison with post-correction values.
Coot correction
A command-line script sends candidate residues and instructions to Coot [9] version 0.5-pre-1 (revision 754) for processing. The residue of interest is first run through real-space refinement, then through the ‘auto-fit rotamer’ tool, followed by a final round of real-space refinement. The first round of real-space refinement optimizes the backbone atoms to the density, allowing for a better chance of correcting the sidechain. Auto-fit rotamer tries each rotamer state defined in Lovell et al. [1] for the given sidechain type and then carries out rigid-body refinement (backbone atoms included). The best-scoring rotamer is accepted. The final round of real-space refinement fine-tunes the atom positions of the selected rotamer to the density, as well as further correcting the position of backbone atoms.
Evaluation of proposed correction
Proposed corrections from Coot are rejected if the rotamer score is below 1%, if the Cβ deviation is >0.25 Å, if the Ramachandran score worsens by more than 30, if the clash overlap worsens by more than 0.01 Å, or if the largest χ angle change is less than 90° (for Leu, Val, or Thr). For Arg, the guanidinium plane is required to change orientation (direction of plane normal) by 180° ± 30°. All accepted changes are large, therefore, and in practice most flip the sidechain over approximately 180° in its density. Proposed changes that pass all these criteria are accepted as valid corrections. Test runs have been done with less stringent criteria and a sample of the output evaluated in order to choose the criteria used here, conservatively considered to ensure producing only genuine improvements (true positives) at the expense of missing some correction possibilities (false negatives).
Output of corrected sidechains
Once each proposed correction has been accepted or rejected, a PDB coordinate file is output incorporating all of the accepted changes. USER MOD records are added to the top of the file summarizing each accepted correction and its quality score evaluations.
Calculation of real-space correlation coefficients
The central procedure in Coot refines real-space correlation coefficients but does not report either initial or final values. For methodological evaluation, therefore, RSCC values for target residues before and after Autofix correction were calculated using the Computational Crystallography Toolbox (CCTBX) [19]. Required mtz files compatible with CCTBX were generated using phenix.refine (PHENIX 1.3-final) with zero macro cycles of refinement.
Results
Large-scale Autofix run
Overall statistics and an example
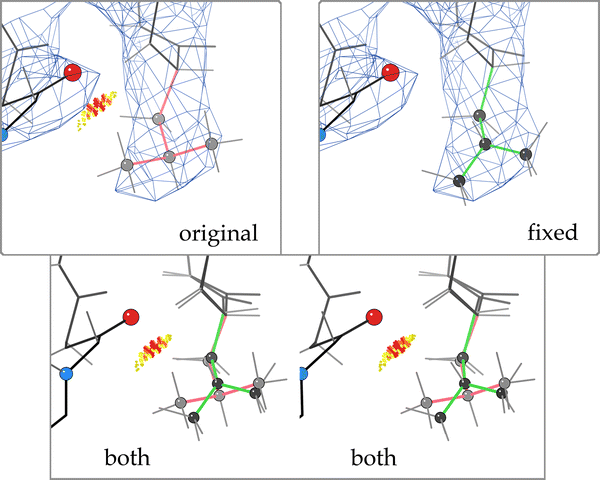
A typical Autofix correction of a backward-fit Leu is illustrated in Fig. 1. The original orientation of Leu D 427 from 1A0E has three validation flags (rotamer outlier, clash, and bad bond angle) but fits the density acceptably. The flipped-over correction uses the density marginally better, with very much improved geometry, sterics, and torsion angles—a clear win all around.
The overall statistics of the Autofix run on Leu, Val, Thr, and Arg sidechain outliers from the 945-file dataset are summarized in Table 1. Overall rates of successful correction (accepted/outliers) are substantial, with Leu (44%), Val (42%), and Thr (32%) performing better than Arg (15%). Leu, Val, and Thr sidechain outliers at high resolution (<2.0 Å) are corrected at a rate of about of 50–70%. Above 2.5 Å resolution, success rates decline, with >3.0 Å structures falling off to only about 15% acceptance rate. For Arg, the more stringent acceptance criteria, plus the limitation of rigid-body refinement in Coot’s auto-fit rotamer step, result in a lower success rate across all resolution ranges. However, the general trend of steep success drop-off at poorer than 3.0 Å resolution is also observed for Arg. Across the board, a total of 3,649 sidechains were automatically corrected in the 945 files, an average of 4 per structure.
Example Autofix correction of a Leu decoy rotamer from the 945-file dataset: Leu D 427 from 1A0E (Thermotoga neapolitana xylose isomerase) at 2.7 Å resolution. a (original) Leu D 427 in its deposited conformation, which is a rotamer outlier with an eclipsed χ angle and a clash with Leu D 430. b (both) Overlay, in stereo, of proposed corrected Leu rotamer (green) over the deposited conformation (pink). c (fixed) Corrected Leu D 427, in a favored mt rotamer. The clash with Leu D 430 has been alleviated and the bond angle idealized, with a somewhat better fit to the density. Images in Figs. 1, 2 and 4 were generated using KING [3]
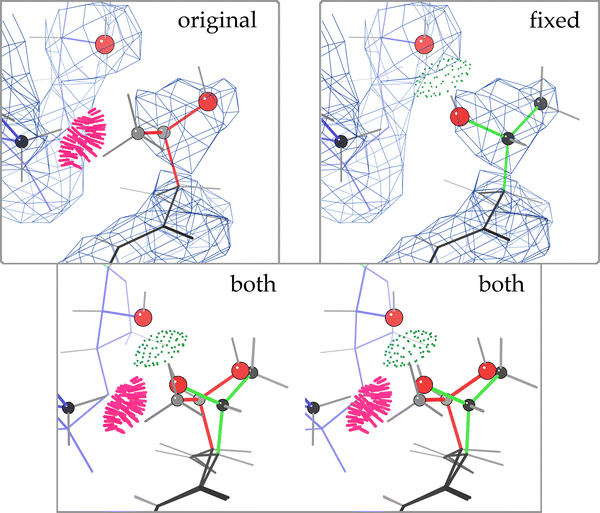
Example Autofix correction from the 50S ribosome: a Thr rotamer outlier, from protein L18e in the 1YHQ archaeal large ribosomal subunit (2.4 Å) [15], before and after correction. a (original) Thr O 3 in its deposited orientation, with fairly good fit to the density, but a serious clash with RNA backbone (Thr methyl to G 0 656 H5′), no H-bond, and a rotamer outlier. b (both) Overlay, in stereo, of proposed corrected Thr rotamer (green) over the original position (pink). c (fixed) Corrected Thr O 3, with equivalent fit to the density, no steric clashes, an excellent p rotamer, and now a strong H-bond from Thr OG1 to the 2′OH of G 0 655. C atoms are gray or black balls; O atoms are larger red balls. Steric clashes are shown as clusters of hot pink spikes, H-bonds as lenses of pale green dots
Test runs on borderline candidates or with looser acceptance criteria
Possibly misfit residues were initially identified in two classes, outlier and borderline (see Methods). Higher acceptance rates are observed for rotamer outliers (score <1%) than for borderline cases (score 1–6%) across all resolution ranges, but most dramatically at high resolution. This is to be expected, as a rotamer score of 6.0% is fairly high and likely to be a correct rotamer already. Examination of individual examples showed that most but definitely not all accepted corrections in the borderline range were valid and worthwhile. As we strive to avoid all erroneous “corrections,” borderline cases are not attempted in the current Autofix protocol, but future enhancements will aim to reliably capture this pool of further sidechain corrections.
Tests were also evaluated with less stringent acceptance criteria in order to optimize the choice of cut-off values. It was immediately evident that rotamer quality should have an absolute cut-off (at the same 1% level used in MolProbity), not just a required improvement. Accepting proposed changes with a small minimum χ angle shift (we tried as low as 10°) was not judged useful, since these cases stay in the same local energy well and will be moved back again by refinement unless explicit hydrogens are included. A 90° shift cut-off was chosen, because it encompasses the two major classes of rotamer errors at tetrahedral branches: the near-180° flips of the decoy rotamers illustrated in the figures, which are the most common systematic error seen in our survey, and also the 120° rotations where there is clear density for only one branch and the wrong alternative rotamer was built. Arg sidechains, with 4 χ angles, are hard to evaluate as well as hard to fit in the first place. Without H-bonds as part of the rotamer refinement (Coot is only using real-space density fit), the requirement of a flipped guanidinium plane was found necessary to prevent proposed changes that were clearly incorrect. Thus, these tests helped us to converge on a relatively simple but stringent set of acceptance criteria.
Autofix therefore tries only outlier candidates and uses the more stringent acceptance criteria described above. Individual examination of a large sample of output found no accepted corrections that were judged incorrect, except when the electron density was very low (usually for an Arg). With initial and final real-space correlation values now available (see below), those cases can be avoided in the future. Although a bad rotamer fit into poor density is quite certainly wrong, its correction would require fitting more than one rotamer alternative, and would be uncomfortably dependent on the non-crystallographic data.
1YHQ ribosomal test case
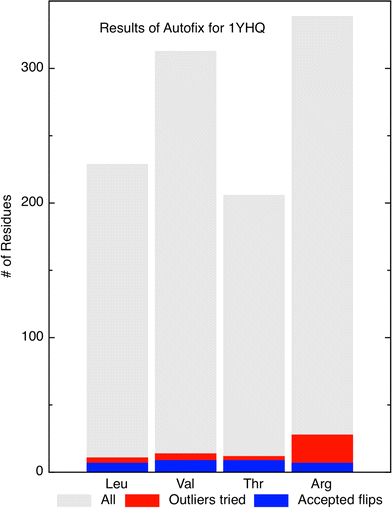
Our Autofix method was also run on the 50S ribosomal subunit structure from Haloarcula marismortui (PDBid: 1YHQ) [15], since we are especially interested in correction of sidechains in protein/RNA contacts. The results for the four residue types are plotted in Fig. 3. Out of 229 total Leu residues, 7 of the 11 outliers were corrected for a success rate of 63%. Out of 313 Val residues, 8 of 14 outliers were corrected for a success rate of 57%. Out of 206 Thr residues, 8 of 12 outliers were corrected for a success rate of 67%. Out of 339 Arg residues, 7 of 28 outliers were corrected for a success rate of 25%. For this large structure, 30 total sidechains were automatically corrected (an average of one per ribosomal protein), many of them in protein/RNA interfaces.
Summary of Autofix results on 1YHQ 50S ribosomal subunit. Bar chart summary of correction results on Leu, Thr, Val, and Arg residues in 1YHQ. Gray bars represent the total number of each residue type in the file. Red represents the number of candidate outliers (<1% rotamer score). Blue represents the number of successfully corrected residues of each type: 7 Leu, 8 Val, 8 Thr, and 7 Arg, which are 63, 57, 67, and 25% of the outliers, respectively
For the 1YHQ 2.4 Å resolution structure, the success rates for correction of outlier candidates is nearly identical to that for typical structures in the 2.0–2.5 Å resolution range (see Table 1) for Leu and Val, somewhat better for Arg (25% vs. 20%) and for Thr (67% vs. 59%). An example of a ribosomal Thr correction at a protein/RNA contact is shown in Fig. 2. Thr O 3 (protein L18e) is corrected to flip a rotamer outlier over into a highly favorable conformation, and to replace a bad steric clash between the Thr methyl and G 0 656 H5′ with a strong H-bond from Thr OG1 to the neighboring ribose O2′. The RSCC decreased from 0.8458 to 0.8340, presumably due to weak and incomplete density for this residue and perhaps some model bias. The backward-fit conformation scores slightly higher because the O atom is further inside the density, but the rotameric fit looks better visually. Coot only used the density to optimize fit, so the excellent angle and distance of the H-bond are an indirect result of identifying the correct local minimum in rotamer space, successfully escaping the initial incorrect local minima.
Leu χ space analysis
Leucine has been previously shown to exhibit two “decoy” rotamer states [1]. These states, denoted mp* and tt*, are rotated 30–40° in χ1 and 140–150° in χ2 from the valid mt and tp rotamers, but their atoms occupy similar regions in space and thus can often fit at least roughly into the same electron density (as in Fig.1). However, the decoys show either internal all-atom clashes or else distorted bond angles to avoid those clashes, have near-eclipsed χ angles, and become less common at lower B-values or higher resolution. In contrast, the related mt and tp states are by far the most populated Leu rotamers and are shown by all validation analyses, and by subsequent refinements [4], to be the correct fitting for these cases.
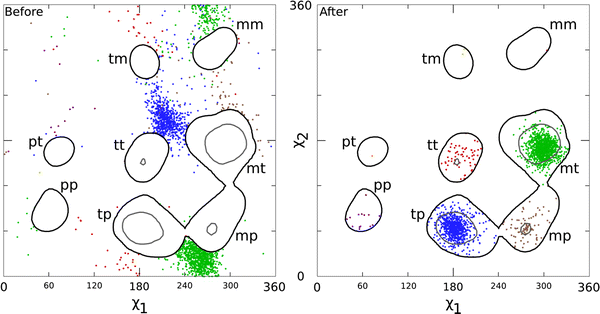
Our analysis identified 4,660 Leu residues in the dataset as rotamer outliers (score <1%), and Autofix corrected 2,037 of these. As shown in Fig. 4 “Before,” these successfully corrected outliers are originally concentrated in the decoy rotamer states (clusters of green and blue data points) and are of course outside the 1% contours. Upon correction (Fig. 4 “After”), the former outliers concentrate highly in the mt and tp rotamers, as expected for correction of such decoys. Outliers that end up in the somewhat less populated rotamers (tt, mp, and pp) are also seen to come from starting conformations clustered nearly 180° away, constituting three new decoy rotamers not described previously.
Before and after χ1–χ2 plots of the 2,037 accepted Leu corrections, for those identified as rotamer outliers (<1%) in our 945-file dataset and successfully corrected by Autofix. Contours are taken from the Top500 Leu set [1], with decoys removed; black lines are the 1% contours and gray lines are the 10% contours of rotamer score. a Before: χ1–χ2 plot for the original conformation of each corrected Leu outlier (thus outside the 1% contours). b After: χ1–χ2 plot of the final χ values for each successfully corrected Leu outlier (now inside the contours). Data points are color-coded by which rotamer they ended up in after correction: mt green, tp blue, tt red, mp brown, pp purple, tm yellow, mm hot pink, and pt orange. Note that for most rotamers, the corrected examples came from a well-defined decoy cluster approximately 180° distant
Real-space correlation validation
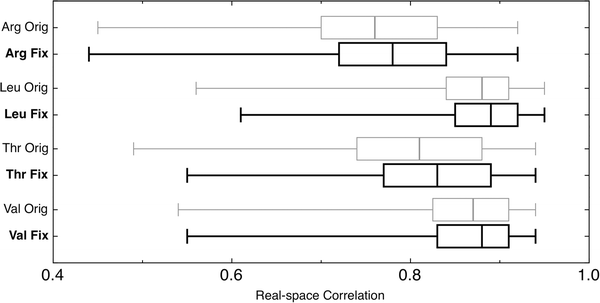
Real-space correlation coefficient (RSCC) calculations were carried out on each corrected residue, both before and after Autofix correction, the outcome of which is summarized in Fig. 5. Median RSCC values and the RSCC range from 25th to 75th percentiles (boxes) improved for all four residue types. One-tailed paired t-tests for each of the residue types are significant at the 99% confidence level and support the hypotheses that the means of the corrected distributions are greater than that of the original distributions, with P-values below 2.2 × 10−16. Leu and Val show the smallest change, since their correlations were already quite high (nearly 0.9) with little room for improvement. The relatively small increases in correlation are expected for two reasons. First, correction of backward-fit residues mostly replaces one heavy atom with another, correcting torsion angles and hydrogen placement to produce little difference in correlation to the data, but a great improvement in chemistry and physics. Second, model bias may limit the increase of RSCC score in some cases. Further refinement would be expected to increase the improvement (see Discussion), and would certainly be done after any production use of Autofix.
Summary of real-space correlation coefficients (RSCC) for corrected outlier residues before (gray) and after (black) Autofix, showing improvement for all 4 amino acid types. Median RSCC values are indicated by a vertical line. The box around the median spans the 25th to the 75th percentile. Whiskers end at the 1st or 99th percentile
Discussion
Conservative correction policy
A major consideration in our development of the Autofix method is ensuring that proposed changes will reliably and robustly be actual improvements. We feel it is acceptable to fail to make all possibly valid corrections (false negatives), but not acceptable to suggest any significant number of changes that either we or the structural biologist would consider clearly wrong upon detailed examination (false positives). Therefore, aspects such as the cut-off levels for acceptance criteria are chosen conservatively. A significant set of proposed corrections for each residue type were visually inspected, which revealed that while outlier candidates were nearly all corrected accurately, many corrections of borderline candidates were dubious and should be rejected. The procedure described here, which only attempts to fix initial rotamer outliers and only accepts results with rotamer score >1%, other scores improved or maintained, and χ angle shifts >90°, does succeed in meeting these goals, while achieving a very useful level of corrections. That is also true for our long-established correction of Asn, Gln, and His sidechain flips in Reduce [2] or in MolProbity [3]. The Autofix methods will gradually be strengthened to cover more cases, as extensions are developed and tested that can do so robustly.
Even with rotamer search, real-space refinement, and stringent requirements for acceptance, a small number of Autofix corrections are found to be false positives. These false positives are seen primarily for Arg residues, from two quite different causes. First, the large hydrogen-bonding capacity of Arginine can sometimes stabilize a sterically unfavorable conformation, which occurs in the starting position of a handful of “corrected” Arg residues (e.g., 1ylt Arg A 256). Neither Coot’s scoring method nor our current acceptance criteria consider H-bonds, and if the starting position is bad enough to be a rotamer outlier then the protocol will be forced to choose some other alternative. Secondly, some surface Arg residues are fit dubiously to weak density, where neither the original nor the corrected residue provides a good answer for a sidechain that almost certainly has multiple conformations.
Additionally, some corrections show a drop rather than in increase in RSCC. This is most often seen for Leu corrections at lower resolution (e.g., 1gpz Leu B 595 at 2.9 Å or 1v4t Leu A 75 at 3.4 Å), where the truncated nubbin of density is best fit by the curled-over, backward-fit conformation, while the correct rotameric fit sticks the CD1/CD2 atoms slightly out of the density. These cases are almost certainly true corrections but cannot be fully substantiated by the sparser, low-resolution data. As density becomes contracted and less clear, Autofix is unable to accurately correct the problem, but such corrections can be done by hand. When a closely related high resolution structure is available it confirms such corrections, such as the nearly 180° backward misfit Leu 68 and 110 of both β chains in the 3.5 Å resolution 2qls hemoglobin, confirmed as standard rotamers in the 1.25 Å 2dn2.
Prevalence of systematic errors
Within our set of 945 PDB files, Table 1 shows that there are a large number of candidate misfit residues for Leu, Thr, Val, and Arg with outlier rotamer scores <1%. For Leu, there are 4,660 candidate outliers, accounting for 8.8% of the total 53,104 Leu residues in the whole set. The 2,037 corrected Leu outliers account for 3.8% of the total Leu residues in the dataset, or on average more than 2 corrected Leu residues per PDB file. A specific example is shown in Fig. 1. While fewer Val and Thr are corrected (1.3% and 1.7%, respectively), there is an average of more than one Val or Thr residue corrected per PDB file. Added to the high rate of Asn/Gln/His flips, this consistent prevalence of rotamer outliers is indicative of a widespread but largely correctable problem in deposited crystal structures. In fact, 99% of the 945 files had at least one Autofix correction. The remaining 1% (10 files) contained no outlier candidates to try correcting. They are all small, high resolution structures: 1b2a (1.7 Å), 1kr0 (1.92 Å), 1w5u (1.14 Å), 1wtf (1.60 Å), 1xyi (1.45 Å), 1ynv (1.2 Å), 1ys0 (2.00 Å), 1zgx (1.13 Å), 2blv (1.2 Å), and 2c9v (1.07 Å).
Resolution effects
The effect of resolution, both on the number of rotamer outliers and on the success of their correction, is an important consideration, documented in Table 1. As resolution decreases, so does the distinct shape of electron density as well as the information content of the diffraction data used in standard crystallographic refinement. Looking at misfit Leu residues, the 364 PDB files with better than 2.0 Å resolutions contain only 497 candidate outliers, for an average of 33 per 1,000 Leu residues. For 2.0–2.5 Å resolution structures, the average jumps to 85 outliers per 1,000 residues; for 2.5–3.0 Å to 112 per 1,000 residues; and for 3.0 Å and poorer all the way to 135 per 1,000 residues. Arg shows a similar pattern. Val and Thr exhibit a lower overall outlier prevalence in this dataset at high resolution, but similar significant increases at poorer than 2.5 Å resolution, with Thr increasing from 80 outlier candidates per 1,000 Thr residues from 2.0 to 2.5 Å resolution to 150 per 1,000 residues >3.0 Å. The ill-defined “blobs” of density at low resolution are less effective for real-space refinement, as they are unable to offer meaningful scoring differences for different proposed rotamer states, so that the ratio of accepted/outlier Autofix corrections goes down. The combined effect of these two trends is that the overall rate of successful corrections is highest in the middle resolution ranges. Interestingly, 52.0% (23,839/45,842) of crystal structures deposited in the PDB as of 10/14/2008 fall within a middle resolution range (1.8–2.5 Å), and the majority of the remaining structures (27.9%: 12,819/45,842) are higher resolution and will have less frequent errors.
Validation by hydrogen-bonding in Thr and Arg
Systematically misfit Thr or Arg residues often have unsatisfied H-bonds and their satisfaction after correction can be taken as an independent validation criterion, since H-bonds were not used in the current protocol. The 1YHQ Thr sidechain shown in Fig. 2 originally had an unsatisfied H-bond as well as a serious clash with the RNA backbone. After correction, it has an equivalent fit to the density, eliminates the clash, and satisfies the H-bond. The guanidinium group at the end of Arg sidechains is also asymmetrical, so that its H-bond interactions are quite different if it is fit flipped-over, producing important disruptions to interactions at molecular interfaces and making its correction an important issue. The examined sample of Arg corrections showed improved H-bonding, often quite dramatically so.
Improvement with refinement
For optimal structure correction, a full round of refinement following Autofix correction is necessary. As shown in Arendall et al. [4], rotameric correction as part of the refinement pipeline improves R and Rfree values and correlation scores. It is important to consider that for that study, corrections and refinement were done in a self-consistent manner, which is limited in this case, as we do not know the complete details of the refinement methods used in each of our dataset structures. We believe that the use of Autofix as part of a self-consistent refinement strategy would yield similar improvements.
Causes of rejected flips
There are a number of reasons that proposed flips are rejected. A primary problem is sidechains with insufficient electron density for valid real-space refinement. In such cases, Coot may either fail to find a changed conformation or may suggest an incorrect rotamer due to an insignificant difference in fit. The latter cases generally but not always produce more all-atom clashes with surrounding groups, larger Cβ deviations, or unfavorable Ramachandran values, so that Autofix can usually correctly reject the proposed change. To ensure that Autofix never accepts a fix without robust real-space evidence, future versions will incorporate a separately calculated real-space correlation value (used but not reported by Coot) as a criterion for acceptance.
A second problem, especially at lower resolution, is other structural errors in the vicinity of the residue of interest. Because Autofix works through candidates one at a time, if a rotamer is corrected but another residue near it is wrong, increased clashes often occur which cause a false rejection of the fix. We cannot accept such changes under our goal of doing no additional harm to the structure, since the false rejections cannot be distinguished from true ones. We plan eventually to treat such interactions combinatorially.
In Fig. 2, note the local backbone movement required to fit the flipped residue into density, which is describable as a “backrub” motion [20]. It is needed because the backward sidechain caused refinement to distort bond angles and shift backbone in order to keep the misfit OG1 and CG2 atoms in density. This example highlights the importance of the two steps of real-space refinement in the Coot component of the Autofix protocol (see Methods), which allowed the necessary motion in this and many cases. For branched-Cβ sidechains in general, even the pre-refinement step does not always improve the direction of the Cα–Cβ bond enough for the correct rotamer to lie in density, so the procedure then fails to identify the flip. Future implementations may therefore incorporate more explicit backrub-type motions.
As a final comment, one should keep in mind that most but not all rotamer outliers are incorrect. About 0.5–1% of sidechains genuinely occupy somewhat strained, outlier conformations (e.g., several hydrogen bonds holding an eclipsed χ angle in a needed position) [1] that are well supported by the electron density and should not be “fixed” by a properly conservative procedure. However, for any pair of atoms that have an all-atom steric clash ≥0.5 Å, one or both of them must be positioned incorrectly. Bond angle outliers >5σ are nearly always incorrect, and are often diagnostic of distortion produced by refinement compensating for groups trapped in the wrong local minimum conformation.
Conclusion and future directions
This initial implementation and testing of the Autofix methodology for correcting backward-fit systematic errors in Leu, Val, Thr, and Arg sidechains has successfully demonstrated that automatic correction is possible for a substantial fraction of the outliers without compromising high reliability of the results. Leu correction shows the highest rate of robust success (44% overall, 69% at high resolution), with Val and Thr close behind. Even with the lower success rate for Arg (15%) those corrections are well-worth making, since the very conservative methodology adopted here ensures that each change is truly an improvement and Arg sidechains make many important long-range interactions. We look forward to such a protocol becoming the standard of good practice in protein crystallography.
In short order, the Autofix methodology will be implemented in MolProbity [3], as an automated correction option available to all users either directly on the web site or by installing a local MolProbity server. The method will also be integrated into the PHENIX system [13, 14], as an intermediary step in the refinement and model completion process to quickly identify these misfit residues and fix them. A similar manual procedure [4] found that such early correction improves refinement behavior as well as accuracy of the final result, which we believe will hold true for the rotameric corrections in Autofix, as well.
As noted in the Discussion, successful correction depends on at least a minimal strength and quality of electron density for the sidechain in question. Future Autofix versions will therefore add real-space correlation value as a criterion for acceptance. The cut-off for acceptance will be chosen after manual evaluation of examples, and may need to be both resolution dependent, and residue type specific, e.g., to account for shortened sidechain density of Leu residues. We will apply Autofix to Ile sidechains, which can also exhibit a systematic flipped state in χ1. We will try out the addition of explicit backrub motions [20]. Addition of an H-bond satisfaction term to real-space refinement of rotamer fits as well as acceptance of corrections should improve future Arg success. Overall, we will study the behavior of modified candidate-selection and acceptance rules in order to expand the number of outlier cases that can be corrected and still maintain high reliability. We plan eventually to treat combinatorially all candidate sidechains that can interact with one another, in a manner similar to the complete H-bond network analysis used in Reduce for hydrogen atom placement and Asn/Gln/His flips [3].
Overall impact
Each individual sidechain correction is a relatively small, local change, but specific atoms move by 2–5 Å. Some examples have quite significant impact on hydrogen-bonding or other specific interactions at active sites, small-molecule binding sites, or protein/nucleic acid interfaces. The result of multiple such corrections lowers crystallographic R and Rfree, improves electron density map interpretability even in other regions, and provides a measurably better protein structure for the end users. In the long run, such methods gradually improve accuracy in the database as a whole, which in turn improves the empirical base behind drug and binding-site design, and improves the accuracy of structural bioinformatics at the atom or residue level, such as motif or H-bonding analysis, internal pairwise preferences used in protein structure validation and prediction, and contact preferences used in the prediction or design of protein/protein and protein/nucleic acid interfaces.
Abbreviations
- Sidechain rotamers are named by their approximate χ angles::
-
p = near +60°, t = trans, m = near −60°
- PDB:
-
Protein Data Bank
References
Lovell SC, Word JM, Richardson JS, Richardson DC (2000) Proteins 40:389–408. doi:10.1002/1097-0134(20000815)40:3<389::AID-PROT50>3.0.CO;2-2
Word JM, Lovell SC, Richardson JS, Richardson DC (1999) J Mol Biol 285:1735–1747. doi:10.1006/jmbi.1998.2401
Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang W, Murray LW, Arendall WBIII, Snoeyink J, Richardson JS, Richardson DC (2007) Nucleic Acids Res 35:W375–W383. doi:10.1093/nar/gkm216
Arendall BWIII, Tempel W, Richardson JS, Zhou W, Wang S, Davis IW, Liu Z-J, Rose JP, Carson WM, Luo M, Richardson DC, Wang B-C (2005) J Struct Funct Genomics 6:1–11. doi:10.1007/s10969-005-3138-4
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) Nucleic Acids Res 28:235–242. doi:10.1093/nar/28.1.235
Vriend G (1990) J Mol Graph 8:52–56. doi:10.1016/0263-7855(90)80070-V
Jones TA, Cowan S, Zou J-Y, Kjeldgaard M (1991) Acta Crystallogr A 47:110–119. doi:10.1107/S0108767390010224
Laskowski RA, Rullmann JA, MacArthur MW, Moss DS, Thornton JM (1993) J Appl Cryst 26:283–291. doi:10.1107/S0021889892009944
Emsley P, Cowtan K (2004) Acta Crystallogr D60:2126–2132. doi:10.1107/S0907444904019158
van den Bedem H, Lotan I, Latombe JC, Deacon AM (2005) Acta Crystallogr D Biol Crystallogr 61(Pt 1):2–13. doi:10.1107/S0907444904025697
Joosten K, Cohen SX, Emsley P, Mooij W, Lamzin VS, Perrakis A (2008) Acta Crystallogr D Biol Crystallogr 64(Pt 4):416–424. doi:10.1107/S0907444908001558
Morris RJ, Perrakis A, Lamzin VS (2003) Methods Enzymol 374:229–244. doi:10.1016/S0076-6879(03)74011-7
Terwilliger TC (2003) Acta Crystallogr D59:38–44. doi:10.1107/S0907444902018036
Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) Acta Crystallogr D58:1948–1954. doi:10.1107/S0907444902016657
Tu D, Blaha G, Moore PB, Steitz TA (2005) Cell 121:257–270. doi:10.1016/j.cell.2005.02.005
Bahadur RP, Zacharias M, Janin J (2008) Nucleic Acids Res 36:2705–2716. doi:10.1093/nar/gkn102
Lovell SC, Davis IW, Arendall BWIII, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC (2003) Proteins 50:437–450. doi:10.1002/prot.10286
Word JM, Lovell SC, LaBean TH, Taylor HC, Zalis ME, Presley BK, Richardson JS, Richardson DC (1999) J Mol Biol 285:1711–1733. doi:10.1006/jmbi.1998.2400
Gross-Kunstleve R (1999) Acta Crystallogr A 55:383–395. doi:10.1107/S0108767398010186
Davis IW, Arendall WBIII, Richardson DC, Richardson JS (2006) Structure 14:265–274. doi:10.1016/j.str.2005.10.007
Acknowledgments
The authors would like to thank Paul Adams, Ralf Grosse-Kunstleve, and Pavel Afonine of Lawrence Berkeley Labs for providing the mtz files necessary for real-space refinement of our dataset and for providing a Python script for automation of the real-space correlation coefficient calculation using the PHENIX CCTBX. Authors are also grateful to our local colleagues Bryan Arendall for help with issues of technical crystallography and with preparation of figures, Vincent Chen for help in interfacing with the quality statistics of MolProbity, Gary Kapral for help with analyzing the 1YHQ ribosome structure, and Lizbeth Videau for initial conversations that inspired this work. This work was supported by National Institutes of Health grants GM063210, GM073919, GM073930, and GM074127, and the Structural Biology and Biophysics Program for DAK.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Headd, J.J., Immormino, R.M., Keedy, D.A. et al. Autofix for backward-fit sidechains: using MolProbity and real-space refinement to put misfits in their place. J Struct Funct Genomics 10, 83–93 (2009). https://doi.org/10.1007/s10969-008-9045-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10969-008-9045-8