Abstract
Toll-like receptors (TLRs) have been shown to participate in the recognition of pathogens by the innate immune system, but it is not clear how a restricted family of receptors has the capacity to recognize the wide spectrum of TLR stimuli known to exist. We report here that two members of the TLR family, TLR2 and TLR6, together coordinate macrophage activation by Gram-positive bacteria and the yeast cell-wall particle, zymosan. TLR6 and TLR2 both are recruited to the macrophage phagosome, where they recognize peptidoglycan, a Gram-positive pathogen component. By contrast, TLR2 recognizes another component, bacterial lipopeptide, without TLR6. The requirement for TLR cooperation is supported by the finding that TLR2 needs a partner to activate tumor necrosis factor-α production in macrophages. Dimerization of the cytoplasmic domain of TLR2 does not induce tumor necrosis factor-α production in macrophages, whereas similar dimerization of the TLR4 cytoplasmic domain does. We show that the cytoplasmic domain of TLR2 can form functional pairs with TLR6 or TLR1, and this interaction leads to cytokine induction. Thus, the cytoplasmic tails of TLRs are not functionally equivalent, with certain TLRs requiring assembly into heteromeric complexes, whereas others are active as homomeric complexes. Finally, we show that TLR6, TLR2, and TLR1 are recruited to macrophage phagosomes that contain IgG-coated erythrocytes that do not display microbial components. The data suggest that TLRs sample the contents of the phagosome independent of the nature of the contents, and can establish a combinatorial repertoire to discriminate among the large number of pathogen-associated molecular patterns found in nature.
Macrophages initiate the innate immune response by recognizing pathogens, phagocytosing them, and secreting inflammatory mediators. An effective response requires that macrophages recognize pathogen-associated molecular patterns (PAMPs) that distinguish the infectious agents from self, and in addition, discriminate among pathogens (1, 2). Two members of the Toll-like receptor (TLR) family have been demonstrated to participate in this process—TLR4 recognizes the Gram-negative product lipopolysaccharide (LPS), whereas TLR2 recognizes various fungal, Gram-positive, and mycobacterial components (3–13). The broad spectrum of components recognized by TLR2, together with the existence of an additional eight TLRs (14–16), suggests that these receptors participate in a complex pattern-recognition system. Each TLR is a type 1 membrane protein, with an extracellular leucine-rich domain that is thought to participate in ligand recognition (17, 18), and an intracellular tail that contains a conserved region called the Toll/IL-1R homology (TIR) domain that, upon activation, results in recruitment of MyD88 (20). MyD88 is an adaptor protein that interacts with the TLRs through its own carboxyl-terminal TIR domain. Through its amino-terminal death domain, MyD88 recruits the serine kinase IRAK to propagate the pro-inflammatory signal (21). Currently, the signaling pathways activated by the TLRs appear to use identical components (20, 22, 23), and it is not clear whether different signals emanate from different members of the TLR family. Here we report that TLR specificity results from cooperation between TLRs. TLR2 and TLR6 are both recruited to the macrophage phagosome, where they recognize peptidoglycan, a Gram-positive pathogen component. TLR2 and TLR6 physically associate, and this interaction leads to cytokine induction. By contrast, TLR2 recognizes another component, bacterial lipopeptide, without TLR6. In addition, we show that the cytoplasmic tails of TLRs are not functionally equivalent and that the cytoplasmic tails of TLR1, TLR2, and TLR6 require partners to activate tumor necrosis factor (TNF)-α production in macrophages, whereas TLR4 is able to do so as a homodimer. The data suggest that TLRs may establish a combinatorial repertoire to discriminate among the large number of PAMPs found in nature.
Materials and Methods
Materials.
The mouse macrophage cell line used in this report, RAW-TT10 (3), is a clone of RAW 264.7 (ATCC no. TIB-71) transfected with an expression vector driving synthesis of the Tet-activator protein, a tetracycline-regulatable transcriptional activator that directs expression, in the absence of tetracycline, from the tetracycline-regulated promoter in the pTIGZ2+ expression vector (3).
DNA Expression Vectors.
Murine TLR6 (GenBank accession no. AF314636) was amplified from RAW 264.7 cDNA by using primers 5′-GAAGAATGGTAAAGTCCC-3′ and 5′-AGTTTTCACATCATCCTC-3′ and was cloned into pEF6/V5-His-TOPO (Invitrogen), which added a V5-epitope tag at the carboxyl terminus (used for immunofluorescence microscopy). Untagged TLR6 was amplified from RAW 264.7 cDNA by using primers 5′-GAAGAATGGTAAAGTCCC-3′ and 5′-TCAAGTTTTCACATCATC-3′ and was cloned into pEF6/V5-His-TOPO [for Chinese hamster ovary (CHO) signaling experiments]. Dominant negative (DN)-TLR6 (P691H) was generated by PCR and confirmed by sequencing. Murine TLR1 (GenBank accession no. AY009154) was cloned by using primers based on the sequence of two murine expressed sequence tags (GenBank accession nos. AA177549 and AA175009). Primer 5′-GCAGCAACATCATTGAGGTGG-3′ was used for reverse transcription of RNA from RAW 264.7 cells, and PCR was performed by using primers 5′-GGCACGTTAGCACTGAGACTC-3′ and 5′-GGTGGATATTCTTATTGCTGTGTG-3′ to yield a 1.8-kb partial sequence. The remaining 5′ sequence was obtained by using 5′ rapid amplification of cDNA ends (RACE; GIBCO/BRL). The full-length TLR1 was hemagglutinin (HA)-tagged at the amino terminus by cloning into pDisplay (Invitrogen), as was described for HA-tagged TLR2, DN-TLR2 (P681H), and DN-TLR4 (P712H) (3). HA-TLR2Δ was generated by PCR to lack the carboxyl-terminal 142 amino acids of TLR2. For single-cell TNF-α assays, DN-TLR2, DN-TLR4, or DN-TLR6 was cloned into the pTIGZ2+ expression vector. pTIGZ2+ uses a tetracycline-regulated promoter (from pTetSplice, GIBCO/BRL) to direct transcription of a bicistronic mRNA that encodes the TLR, followed by a cap-independent translational enhancer region (from pCITE, Novagen) that drives translation of enhanced green fluorescent protein (GFP; CLONTECH; ref. 3).
Transfection.
RAW-TT10 macrophages and CHO-K1 (ATCC no. CRL-9618) cells were transfected by using a method described in ref. 4. All experiments were performed 24 h after transfection.
Detection of Intracellular TNF-α.
Clinical isolates of Staphylococcus aureus, Streptococcus pneumoniae, and Enterococcus were obtained from the University of Washington Medical Center, Seattle. Listeria monocytogenes (strain 10403S) was obtained from D. Portnoy (University of California, Berkeley), Lactobacillus was provided by S. Klebanoff (University of Washington, Seattle), and Salmonella minnesota was from the American Type Culture Collection (no. 49284). Bacteria were grown to saturation, heat killed, and stored as frozen aliquots. For particle stimulation, the RAW macrophages were incubated for 30 min at 37°C with 3 × 106 zymosan (Molecular Probes), Staphylococcus aureus, or Salmonella minnesota particles per well in 24-well plates. After exposure to the particles, the cells were incubated for 4 h at 37°C in the presence of 5 μg/ml brefeldin A to accumulate intracellular TNF-α. Where indicated, cells were exposed to 10 μg/ml peptidoglycan (Staphylococcus aureus, Fluka), 100 ng/ml Pam3CSK4 (Roche), or 10 ng/ml LPS (Salmonella minnesota R595, List, Campbell, CA; identical results were obtained with Escherichia coli LPS O111:B4 and O55:B5), together with 5 μg/ml brefeldin A for 4 h at 37°C. After blocking Fc receptors with 2.4G2 hybridoma supernatant, the cells were fixed with 4% (vol/vol) paraformaldehyde in PBS. The cells were permeabilized and stained with phycoerythrin-conjugated anti-mouse TNF-α antibody (PharMingen), diluted in 1% FCS and 0.1% saponin in PBS. After two washes, the cells were analyzed by flow cytometry by using a FACScan (Becton Dickinson), and WinMDI software (Joseph Trotter, Scripps Research Institute, La Jolla, CA). By using GFP expression as an indicator of transgene expression, TNF-α produced in transfected cells expressing DN-TLR could be compared directly with cells in the same sample that failed to express DN-TLR.
Immunofluorescence Microscopy.
RAW-TT10 macrophages were transfected with V5-tagged TLR6, HA-tagged TLR1, or HA-tagged TLR2, and grown overnight on glass coverslips. After incubation with zymosan (Molecular Probes) or IgG-opsonized sheep red blood cells (prepared as described in ref. 24) for 5 min at 37°C, the cells were fixed and permeabilized, and nonspecific sites were blocked by incubation in blocking buffer (PBS/10% calf serum/0.1% ovalbumin/5 mM sodium azide). Epitope-tagged TLRs were detected by using mouse monoclonal antibody to the V5 epitope (Invitrogen) or the HA epitope (HA.11, Covance, Princeton, NJ). Specific binding was detected with fluorescein-conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch). After mounting, the cells were observed by fluorescence microscopy (Axiovert 100TV microscope, Zeiss) using an MRC 1024 confocal microscope (Bio-Rad).
Immunoprecipitation.
CHO-K1 cells were transiently transfected with 5 μg of TLR2 (HA-epitope-tagged), TLR6 (V5-epitope-tagged), CD4-TLR1, CD4-TLR2, CD4-TLR4, or CD4-TLR6 as indicated. Twenty-four hours after transfection, cells were stimulated, where indicated, with peptidoglycan (10 μg/ml for 10 min). After lysis (1% Triton X-100/10 mM Hepes, pH 7.4/150 mM NaCl) and centrifugation (18,000 × g, for 10 min), aliquots of the lysates were retained for Western blotting to confirm equivalent expression of HA-TLR2 or CD-TLR chimeras, as indicated. Lysates were precleared (for 30 min) with rabbit antibody coupled to Sepharose beads, and TLR6 was immunoprecipitated with anti-V5 antibody (Invitrogen) and Sepharose beads (Gammabind, Amersham Pharmacia). After washing, immunoprecipitated proteins were eluted by boiling in SDS/PAGE loading buffer, separated by 7% (vol/vol) SDS/PAGE (nonreducing conditions were used when followed by CD4 detection), and transferred to poly(vinylidene difluoride) membrane. Coimmunoprecipitated HA-TLR2 was identified with anti-HA antibody (HA.11, Covance), and the CD4-TLR chimeras were identified with anti-CD4 (PharMingen). The primary antibodies were detected by chemiluminescence (Lumi-light, Roche) with horseradish peroxidase-conjugated anti-mouse antibody (Zymed).
Luciferase Assays.
Activation of NF-κB by TLRs was measured as described (3, 4). CHO-K1 cells were transiently transfected with 2 μg of an NF-κB reporter construct (ELAM-1 firefly luciferase; ref. 25) and 0.2 μg of a construct directing expression of Renilla luciferase under the control of the constitutively active thymidine kinase promoter (pRL-TK, Promega), together with 1 μg of mCD14 expression vector and 3 μg of TLR expression constructs (3 μg for one TLR, 1.5 μg each for two TLRs), as indicated in the text, and plated into 24-well tissue culture plates. Twenty-four hours after transfection, cells were stimulated, where indicated, with 10 μg/ml peptidoglycan or 100 ng/ml Pam3CSK4 lipopeptide for 4 h, and luciferase activity was measured by using the Dual-Luciferase Reporter Assay System (Promega), according to the manufacturer's instructions. Data are presented as the mean ± standard deviation of triplicate samples and are representative of three independent experiments.
Constitutively Active CD4-TLRs.
Constitutively active TLRs were constructed by fusing cDNAs encoding the extracellular domain of murine CD4 (amino acids 1–391) to the transmembrane and cytoplasmic domains of murine TLR1 (amino acids 586–795), TLR2 (amino acids 590–784), TLR4 (amino acids 628–835), and TLR6 (amino acids 598–806). The constructs were cloned into the pEF6/V5-His-TOPO (Invitrogen) expression vector. RAW-TT10 macrophages were transiently transfected with the indicated CD4-TLR constructs (10 μg for one vector or 5 μg each for two vectors). After 18 h, 5 μg/ml brefeldin A was added (for 4 h) to accumulate intracellular TNF-α, which was measured by intracellular cytokine staining, as described above. FITC-anti-CD4 (PharMingen) was used to detect transfected cells. Gates were set to compare TNF-α production in cells expressing identical levels of the CD4-TLR constructs. Data are shown from a single experiment, and are representative of three independent experiments.
Results and Discussion
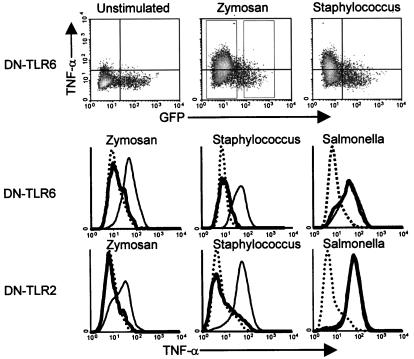
An understanding of the role for TLRs in PAMP recognition by macrophages has been facilitated by the use of DN receptors. A natural DN mutation in TLR4 underlies LPS insensitivity in mice (19, 26, 27), whereas a similar mutation in TLR2 suppresses responses to cell wall components of Gram-positive bacteria, mycobacteria, and fungi (3, 4). TLR6 is related to both TLR2 and TLR4, although nothing is known about its ligands (28). We found that mutation of proline-691 to histidine generated a dominant negative form of TLR6 (DN-TLR6), as occurs when the homologous prolines are mutated in TLR2 and TLR4 (3, 29). RAW-TT10 mouse macrophages were transiently transfected with an expression vector encoding a bicistronic mRNA transcript that directed expression of both DN-TLR6 and GFP (3). The cells were stimulated with yeast cell wall particles (zymosan) or bacteria, and the production of TNF-α was measured by intracellular staining and flow cytometry. The expression of GFP and DN-TLR6 from the same mRNA transcript permitted a direct comparison of TNF-α production in DN-TLR6-expressing cells (GFP-positive) with cells in the same sample that did not express the inhibitory protein. Thus, cells that were stimulated with zymosan, and that did not express the inhibitory protein (GFP-negative cells), produced TNF-α (Fig. 1). By contrast, DN-TLR6 inhibited zymosan-stimulated TNF-α production to background levels (Fig. 1). DN-TLR6 also completely inhibited TNF-α production stimulated by the Gram-positive bacterium Staphylococcus aureus, whereas it had no effect on TNF-α production induced by the Gram-negative bacterium Salmonella minnesota. The spectrum of pathogens recognized by TLR6 mirrored the specificity of TLR2 (Fig. 1; ref. 3). In addition, expression of DN-TLR6 or DN-TLR2 completely inhibited TNF-α production elicited by other Gram-positive bacteria, including Streptococcus pneumoniae, Listeria monocytogenes, Lactobacillus crispatus, and Enterococcus faecalis (data not shown). By contrast, expression of DN-TLR4 solely inhibited TNF-α production elicited by Gram-negative bacteria, and had no effect on responses to Gram-positive bacteria (data not shown). The observation that DN-TLR6 and DN-TLR2 independently and completely inhibit TNF-α production elicited by whole Gram-positive bacteria suggests that together, these two receptors coordinate this proinflammatory signaling.
Figure 1.
TLR2 and TLR6 are required for the macrophage TNF-α response to yeast and Gram-positive bacteria. Macrophages transfected with DN-TLR6 or DN-TLR2 were stimulated with the indicated microbes, and the production of TNF-α was measured by flow cytometry. The density plots (Top) correlate the level of DN-TLR expression (GFP, x axis) with the production of TNF-α (y axis). By using the gates indicated in the zymosan-stimulated density plot (dotted rectangles), histograms were generated for TNF-α production by DN-TLR- expressing and nonexpressing cells. Thin lines indicate TNF-α production by macrophages that did not express DN-TLRs. Thick lines indicate TNF-α production by cells expressing DN-TLR6 (Middle) or DN-TLR2 (Bottom). Dotted lines indicate the background level of TNF-α production in unstimulated cells. The y axes indicate relative cell number.
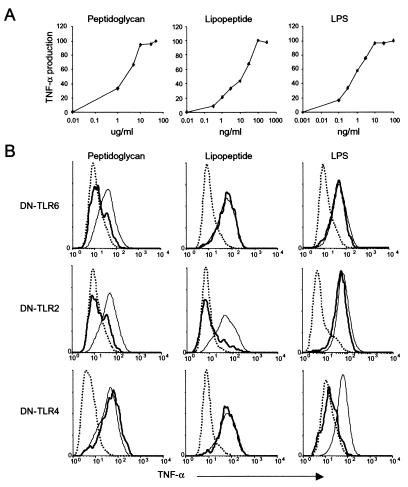
To test whether TLR6 and TLR2 signaling is initiated by identical PAMPs, we stimulated macrophages with the purified bacterial cell wall components, peptidoglycan, the generic synthetic tripalmitoylated lipopeptide (Pam3CSK4), and LPS (Fig. 2). To exclude the effect of differences in the sensitivity of the macrophages to the different stimuli, we stimulated the macrophages with the lowest dose that elicited a maximal TNF-α response (Fig. 2A). Peptidoglycan (10 μg/ml), a major surface component of Gram-positive bacteria, strongly induced TNF-α in macrophages (Fig. 2B). Expression of either DN-TLR6 or DN-TLR2 completely inhibited peptidoglycan-induced TNF-α production (Fig. 2B). However, the PAMP specificity of TLR6 and TLR2 was not completely overlapping. Bacterial lipoproteins are a family of proinflammatory cell wall components found in both Gram-positive and Gram-negative bacteria (11, 30). The stimulatory activity of a bacterial lipoprotein resides in its acylated amino terminus, and is mimicked by the Pam3CSK4 lipopeptide, which recently has been shown to activate the proinflammatory transcription factor, NF-κB, in a TLR2-dependent manner (8, 9, 11). Pam3CSK4 (100 ng/ml) stimulated TNF-α production in macrophages, and expression of DN-TLR2 completely inhibited this response (Fig. 2B). By contrast, expression (over a concentration range of 103) of DN-TLR6 had no effect on Pam3CSK4-induced TNF-α production (Fig. 2B and data not shown). DN-TLR6 also had no effect on TNF-α production induced by lower doses of Pam3CSK4 (half-maximal stimulatory dose = 14 ng/ml; Fig. 2A). In addition, DN-TLR6 had no effect on Gram-negative bacterial LPS-induced TNF-α production, which was mediated by TLR4 (Fig. 2B). Together, these results demonstrate that TLR2, TLR4, and TLR6 each have a defined ligand specificity, with the specificities of TLR6 and TLR2 partially overlapping. Furthermore, the results with Pam3CSK4 and LPS confirm the specificity of the assay, and exclude the possibility that the effect of DN-TLR6 is to inhibit the activity of TLR2 or TLR4, respectively.
Figure 2.
Macrophages use TLR2, TLR4, and TLR6 to induce the production of TNF-α in response to different microbial components. (A) Mock-transfected RAW-TT10 macrophages were stimulated with the indicated doses of peptidoglycan, Pam3CSK4 lipopeptide, or LPS, and the production of TNF-α was measured by flow cytometry. The y axis indicates the percent maximal TNF-α production for each stimulus. Maximal TNF-α production was induced by 10 μg/ml peptidoglycan, 100 ng/ml Pam3CSK4 lipopeptide, or 10 ng/ml LPS, and these doses were used in B. (B) Macrophages transfected with dominant negative forms of TLR2, TLR4, or TLR6 were stimulated with the indicated microbial components, and the production of TNF-α was measured by flow cytometry. Thin lines indicate TNF-α production by macrophages that did not express DN-TLRs. Thick lines indicate TNF-α production by cells expressing DN-TLR6 (Top), DN-TLR2 (Middle), or DN-TLR4 (Bottom). Dotted lines indicate the background level of TNF-α production in unstimulated cells. The y axes indicate relative cell number.
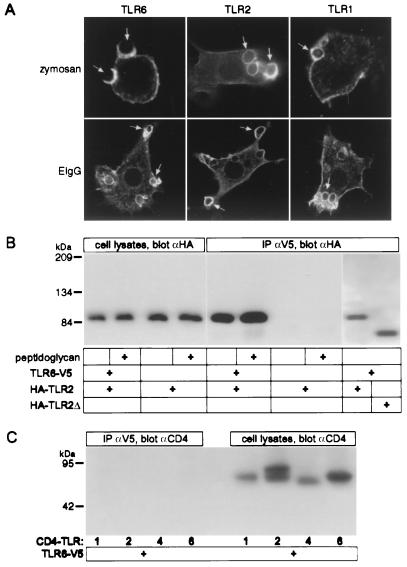
The phagocytosis of particles is coupled to the production of inflammatory mediators. Like TLR2, TLR6 is expressed at the cell surface and is enriched on phagosomes containing zymosan (Fig. 3A; ref. 3). Interestingly, TLR2 and TLR6 both are recruited to phagosomes that contain additional PAMPs such as Gram-positive and Gram-negative bacteria (data not shown), as well as to phagosomes that do not contain PAMPs, such as those containing IgG-opsonized red blood cells (Fig. 3A). This suggests that TLRs are recruited to all phagosomes where they can sample the contents and potentially identify the nature of the pathogen. This would imply that additional TLRs also are recruited to phagosomes, a hypothesis that was supported by the observation that TLR1 was enriched on phagosomes containing either zymosan or IgG-opsonized sheep red blood cells (Fig. 3A).
Figure 3.
TLR6 is enriched on macrophage phagosomes and physically associates with TLR2. (A) V5-epitope-tagged TLR6, HA-epitope-tagged TLR2, and HA-epitope-tagged TLR1 were localized by immunofluorescence microscopy in transiently transfected RAW macrophages after the phagocytosis of zymosan or IgG-coated sheep erythrocytes [EIgG (arrows)] for 5 min. (B) CHO cells were transfected with HA-epitope-tagged TLR2 with or without V5-epitope-tagged TLR6 and were stimulated, where indicated, with peptidoglycan (10 μg/ml for 10 min). TLR2 was detected by Western blotting in the cell lysates, and by co-immunoprecipitation (IP) with TLR6. A truncated form of TLR2 (lacking a cytoplasmic tail, HA-TLR2Δ) also was coimmunoprecipitated with TLR6. (C) CHO cells were transfected with V5-epitope-tagged TLR6 and chimeric receptors composed of the extracellular domain of CD4 fused to the transmembrane domain and the cytoplasmic tail of TLR1 (CD4-TLR1), TLR2 (CD4-TLR2), TLR4 (CD4-TLR4), or TLR6 (CD4-TLR6). The CD4-TLR chimeras were detected by Western blotting in the cell lysates, but did not coimmunoprecipitate with TLR6.
Next, we examined whether TLR2 and TLR6 physically interact. CHO cells were cotransfected with epitope-tagged TLR6 and TLR2. Immunoprecipitation of V5-tagged TLR6 resulted in coprecipitation of HA-tagged TLR2, indicating that they were physically associated (Fig. 3B). The extent of the association between TLR6 and TLR2 was not influenced by the addition of stimulatory bacterial components (Fig. 3B). The association of TLR2 and TLR6 was mediated by their extracellular domains, because deletion of their cytoplasmic domains did not prevent their interaction (Fig. 3B and data not shown). Conversely, deleting the extracellular domain abrogated the association of TLRs. When the cytoplasmic and transmembrane domains of TLR1, -2, -4, or -6 were fused to the extracellular domain of CD4, they did not associate with wild-type TLR6 (Fig. 3C). Thus, in CHO cells, TLR2 and TLR6 readily associate with each other in a ligand-independent manner through their extracellular domains.
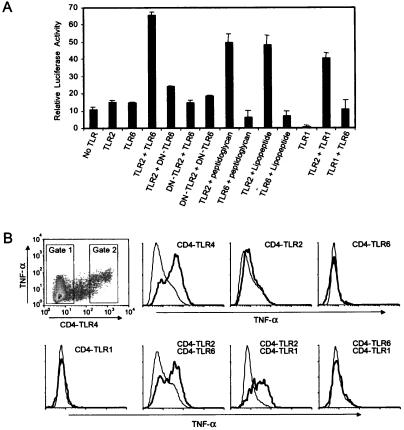
The observation that TLR2 and TLR6 were constitutively associated led us to examine their signaling capacity in CHO cells. Expression of either TLR2 or TLR6 did not lead to the activation of NF-κB, a transcription factor required for proinflammatory signaling (Fig. 4A). However, coexpression of TLR2 and TLR6 strongly activated NF-κB (Fig. 4A), under conditions in which these molecules physically associate (Fig. 3B). This ligand-independent constitutive activity required functional TLR2 and TLR6, because expression of either DN-TLR2 or DN-TLR6 did not result in NF-κB activation (Fig. 4A). In addition, these data demonstrate that DN-TLR6 is a signaling-deficient receptor, as was described for DN-TLR2 and DN-TLR4 (3, 29). Because coexpressed TLR2 and TLR6 were constitutively associated and active in CHO cells, we were unable to study the effects of ligands on TLR association or signaling. Golenbock and coworkers have demonstrated that CHO cells are genetically deficient in TLR2 (31), and they and others also have shown that ectopic expression of TLR2 in these cells permits reconstitution of signaling induced by peptidoglycan and lipopeptide (8, 9, 11, 12). We obtained identical results (Fig. 4A). However, expression of TLR6 in CHO cells failed to reconstitute peptidoglycan-induced signaling (Fig. 4A), further supporting a requirement for TLR2 in TLR6-mediated recognition of this PAMP. As expected, TLR6 also did not reconstitute bacterial lipopeptide-induced signaling in CHO cells (Fig. 4A).
Figure 4.
TLR6 cooperates with TLR2 in mediating proinflammatory signals. (A) The relative activity of an NF-κB luciferase reporter was assessed in CHO cells transiently expressing the indicated TLR constructs without further stimulation, or after stimulation by the indicated bacterial products. (B) RAW-TT10 macrophages were transfected with the indicated CD4-TLR chimeric constructs, and the production of TNF-α was measured by flow cytometry. The representative density plot shows that expression of CD4-TLR4 results in the production of TNF-α. Histograms were generated by using the gates shown. Thin lines indicate TNF-α production by macrophages that did not express the indicated CD4-TLRs (gate 1). Thick lines indicate TNF-α production by cells expressing CD4-TLRs (gate 2). The y axes of the histograms indicate relative cell number, and the x axes indicate TNF-α production on a log scale identical to that shown (on the y axis) in the density plot.
When paired, TLR2 and TLR6 activate NF-κB; alone, however, these receptors do not signal (Fig. 4A). Because TLR2 is capable of signaling in a TLR6-independent manner when stimulated with lipopeptide (Fig. 2), we examined whether it formed a signaling complex with another TLR. Coexpression of TLR1 with TLR2 resulted in robust activation of the NF-κB pathway, whereas TLR1, alone, did not (Fig. 4A). Formation of this functional complex was specific, because coexpression of TLR1 and TLR6 did not signal. Thus, certain combinations of TLRs appear to form functional signaling complexes. This hypothesis was further examined in RAW macrophages, where production of an inflammatory cytokine, TNF-α, can be measured.
The extracellular domain of CD4 is known to promote homodimerization of the molecule (32, 33). Chimeras composed of the extracellular domain of CD4 fused with the transmembrane and intracellular region of TLR4 are constitutively active (6, 34). We generated chimeras in which the extracellular domains of TLR1, TLR2, TLR4, or TLR6 were replaced by the corresponding region of CD4, and we examined their signaling properties in macrophages. As reported (34), CD4-TLR4 induced TNF-α production in macrophages (Fig. 4B). By contrast, neither CD4-TLR1, nor CD4-TLR2, nor CD4-TLR6 induced TNF-α production (Fig. 4B), even though they were expressed at identical levels to CD4-TLR4 on the macrophage surface. These data indicate that the signaling domains of TLR1, TLR2, and TLR6 are not functionally equivalent to that of TLR4. The finding that CD4-TLR2 homodimers are not active suggests that TLR2 needs a signaling partner to activate TNF-α production. When CD4-TLR2 and CD4-TLR6 were coexpressed, TNF-α production was induced (Fig. 4B). Similarly, coexpression of CD4-TLR2 and CD4-TLR1 induced robust production of TNF-α (Fig. 4B). However, there was specificity in this interaction, because coexpression of CD4-TLR1 and CD4-TLR6 did not induce cytokine production (Fig. 4B). Thus, as seen in the CHO system, TLR2 can pair with either TLR6 or TLR1 to establish a functional signaling complex.
We have demonstrated that TLR6 cooperates with TLR2 in identifying a variety of PAMPs. In macrophages, expression of either DN-TLR2 or DN-TLR6 completely inhibited TNF-α production elicited by Gram-positive bacteria or yeast, and both receptors were recruited to phagosomes. In CHO cells, TLR2 physically associated with TLR6, leading to the constitutive activation of NF-κB. In macrophages, coexpression of CD4-TLR2 and CD4-TLR6 activated TNF-α production, whereas expression of either receptor alone was insufficient to induce TNF-α production. TLR2 and TLR6 cooperate in detecting bacterial components such as peptidoglycan, but they do not have identical specificity, as TLR2 detects bacterial lipopeptide without support from TLR6. This latter finding implicates another, as yet unidentified, TLR as a partner for TLR2 in the detection of bacterial lipopeptide, because our data indicate that CD4-TLR2 homodimers are not sufficient to activate TNF-α production in macrophages. We show that TLR2 is capable of forming a functional signaling complex with TLR1, indicating that TLR2 has multiple partners with which it can combine. However, we have not been able to demonstrate that TLR1/TLR2 complexes mediate the detection of lipopeptide (data not shown), suggesting that TLR2 has yet additional partners.
TLR2 has been shown to detect a wide array of PAMPs, and it has been unclear how a single molecule could accommodate such diverse ligands. Our data indicate that TLR2 forms functional complexes with TLR6, and perhaps with other TLRs, and thereby increases its range of detection. It is not yet clear whether TLRs recognize PAMPs directly, as suggested by recent data with TLR4 (19), or through endogenous ligands generated in response to PAMPs (35).
It is particularly interesting that the highly homologous cytoplasmic domains of TLRs are not functionally equivalent with respect to signaling. For example, whereas dimerization of the TLR4 cytoplasmic domain stimulates TNF-α production, similar dimerization of the cytoplasmic domains of TLRs 1, 2, and 6 does not. The cytoplasmic domains of TLR1 and TLR6, however, can form functional pairs with TLR2. One possible implication of this combinatorial diversity is that different TLR pairs may stimulate different signaling pathways. Such differential signaling would allow macrophages to tailor their responses to individual pathogens by producing specific patterns of inflammatory mediators.
The innate immune system has to recognize the large number of PAMPs found in nature. It must both distinguish these structures from self, and discriminate between different pathogens to mount an appropriate immune defense. To date, 10 human TLRs have been identified. Our data suggest that these molecules might form a combinatorial repertoire for innate immune recognition.
Acknowledgments
We thank members of the Department of Immunology for advice with the manuscript. This work was supported in part by grants from the National Institutes of Health, AI25032 (to A.A.), AI32972 (to A.A.), HL65898 (to C.B.W.), and from the Cystic Fibrosis Foundation (to C.B.W. and A.M.H.).
Abbreviations
- TLR
Toll-like receptor
- TNF
tumor necrosis factor
- PAMP
pathogen-associated molecular pattern
- LPS
lipopolysaccharide
- DN
dominant negative
- HA
hemagglutinin
- GFP
green fluorescent protein
- Pam
palmitoyl
- CHO
Chinese hamster ovary
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF314636 and AY009154).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250476497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250476497
References
- 1.Janeway C A., Jr Cold Spring Harbor Symp Quant Biol. 1989;54:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Janeway C A., Jr Immunol Today. 1992;13:11–16. doi: 10.1016/0167-5699(92)90198-G. [DOI] [PubMed] [Google Scholar]
- 3.Underhill D M, Ozinsky A, Hajjar A M, Stevens A, Wilson C B, Bassetti M, Aderem A. Nature (London) 1999;401:811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- 4.Underhill D M, Ozinsky A, Smith K D, Aderem A. Proc Natl Acad Sci USA. 1999;96:14459–14463. doi: 10.1073/pnas.96.25.14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 6.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 7.Takeuchi O, Takeda K, Hoshino K, Adachi O, Ogawa T, Akira S. Int Immunol. 2000;12:113–117. doi: 10.1093/intimm/12.1.113. [DOI] [PubMed] [Google Scholar]
- 8.Brightbill H D, Libraty D H, Krutzik S R, Yang R B, Belisle J T, Bleharski J R, Maitland M, Norgard M V, Plevy S E, Smale S T, et al. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 9.Lien E, Sellati T J, Yoshimura A, Flo T H, Rawadi G, Finberg R W, Carroll J D, Espevik T, Ingalls R R, Radolf J D, Golenbock D T. J Biol Chem. 1999;274:33419–33425. doi: 10.1074/jbc.274.47.33419. [DOI] [PubMed] [Google Scholar]
- 10.Means T K, Wang S, Lien E, Yoshimura A, Golenbock D T, Fenton M J. J Immunol. 1999;163:3920–3927. [PubMed] [Google Scholar]
- 11.Aliprantis A O, Yang R B, Mark M R, Suggett S, Devaux B, Radolf J D, Klimpel G R, Godowski P, Zychlinsky A. Science. 1999;285:736–739. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- 12.Yoshimura A, Lien E, Ingalls R R, Tuomanen E, Dziarski R, Golenbock D. J Immunol. 1999;163:1–5. [PubMed] [Google Scholar]
- 13.Flo T H, Halaas O, Lien E, Ryan L, Teti G, Golenbock D T, Sundan A, Espevik T. J Immunol. 2000;164:2064–2069. doi: 10.4049/jimmunol.164.4.2064. [DOI] [PubMed] [Google Scholar]
- 14.Rock F L, Hardiman G, Timans J C, Kastelein R A, Bazan J F. Proc Natl Acad Sci USA. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaudhary P M, Ferguson C, Nguyen V, Nguyen O, Massa H F, Eby M, Jasmin A, Trask B J, Hood L, Nelson P S. Blood. 1998;91:4020–4027. [PubMed] [Google Scholar]
- 16.Hardiman, G. T., Rock, F. L., Bazan, J. F. & Kastelein, R. A. (1998) World Intellectual Property Organization Patent WO 98/50547.
- 17.Goddard, A., Godowski, P. J., Gurney, A. L., Mark, M. A. & Yang, R-B. (1999) World Intellectual Property Organization Patent WO 99/20756.
- 18.Lien E, Means T K, Heine H, Yoshimura A, Kusumoto S, Fukase K, Fenton M J, Oikawa M, Qureshi N, Monks B, Finberg R W, et al. J Clin Invest. 2000;105:497–504. doi: 10.1172/JCI8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poltorak A, Ricciardi-Castagnoli P, Citterio S, Beutler B. Proc Natl Acad Sci USA. 2000;97:2163–2167. doi: 10.1073/pnas.040565397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway C A., Jr Mol Cell. 1998;2:253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 21.Muzio M, Ni J, Feng P, Dixit V M. Science. 1997;278:1612–1615. doi: 10.1126/science.278.5343.1612. [DOI] [PubMed] [Google Scholar]
- 22.Muzio M, Natoli G, Saccani S, Levrero M, Mantovani A. J Exp Med. 1998;187:2097–2101. doi: 10.1084/jem.187.12.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F X, Kirschning C J, Mancinelli R, Xu X P, Jin Y, Faure E, Mantovani A, Rothe M, Muzio M, Arditi M. J Biol Chem. 1999;274:7611–7614. doi: 10.1074/jbc.274.12.7611. [DOI] [PubMed] [Google Scholar]
- 24.Gold E S, Morrissette N S, Underhill D M, Guo J, Bassetti M, Aderem A. Immunity. 2000;12:285–292. doi: 10.1016/s1074-7613(00)80181-8. [DOI] [PubMed] [Google Scholar]
- 25.Schindler U, Baichwal V R. Mol Cell Biol. 1994;14:5820–5831. doi: 10.1128/mcb.14.9.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poltorak A, He X, Smirnova I, Liu M Y, Huffel C V, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 27.Qureshi S T, Lariviere L, Leveque G, Clermont S, Moore K J, Gros P, Malo D. J Exp Med. 1999;189:615–625. doi: 10.1084/jem.189.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeuchi O, Kawai T, Sanjo H, Copeland N G, Gilbert D J, Jenkins N A, Takeda K, Akira S. Gene. 1999;231:59–65. doi: 10.1016/s0378-1119(99)00098-0. [DOI] [PubMed] [Google Scholar]
- 29.Du X, Poltorak A, Silva M, Beutler B. Blood Cells Mol Dis. 1999;25:328–338. doi: 10.1006/bcmd.1999.0262. [DOI] [PubMed] [Google Scholar]
- 30.Janeway C A, Jr, Medzhitov R. Curr Biol. 1999;9:R879–82. doi: 10.1016/s0960-9822(00)80073-1. [DOI] [PubMed] [Google Scholar]
- 31.Heine H, Kirschning C J, Lien E, Monks B G, Rothe M, Golenbock D T. J Immunol. 1999;162:6971–6975. [PubMed] [Google Scholar]
- 32.Koskinen R, Lamminmaki U, Tregaskes C A, Salomonsen J, Young J R, Vainio O. J Immunol. 1999;162:4115–4121. [PubMed] [Google Scholar]
- 33.Wu H, Kwong P D, Hendrickson W A. Nature (London) 1997;387:527–530. doi: 10.1038/387527a0. [DOI] [PubMed] [Google Scholar]
- 34.Medzhitov R, Preston-Hurlburt P, Janeway C A., Jr Nature (London) 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 35.Medzhitov R, Janeway C A., Jr Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]