Abstract
Recent experimental studies of the denatured state and theoretical analyses of the folding landscape suggest that there are a large multiplicity of low-energy, partially folded conformations near the native state. In this report, we describe a strategy for predicting protein structure based on the working hypothesis that there are a greater number of low-energy conformations surrounding the correct fold than there are surrounding low-energy incorrect folds. To test this idea, 12 ensembles of 500 to 1,000 low-energy structures for 10 small proteins were analyzed by calculating the rms deviation of the Cα coordinates between each conformation and every other conformation in the ensemble. In all 12 cases, the conformation with the greatest number of conformations within 4-Å rms deviation was closer to the native structure than were the majority of conformations in the ensemble, and in most cases it was among the closest 1 to 5%. These results suggest that, to fold efficiently and retain robustness to changes in amino acid sequence, proteins may have evolved a native structure situated within a broad basin of low-energy conformations, a feature which could facilitate the prediction of protein structure at low resolution.
Prediction of the structures of proteins from their amino acid sequence traditionally has followed one approach. First, a candidate conformation is generated, either by a de novo conformational search method or by turning to the database of known protein structures. This conformation then is scored for the quality of the match between the sequence of the target protein and the spatial positions forced on the residues when placed in the candidate conformation. This process is continued until practical limitations force termination of the search, at which point the conformation with the most favorable score is considered to be the best candidate for the structure of the target protein.
A central assumption underlying this standard approach is that the native state is the conformation of lowest energy. The configurational entropy of the protein chain cannot be included in the scoring function because the focus is on finding one conformation. Consequently, this approach is not rigorously based on Anfinsen’s hypothesis that the native state lies at the global minimum in free energy (1). Interpreted literally, the Anfinsen hypothesis implies that, because the native state of a protein is an ensemble of many similar conformations, the target or goal of protein structure prediction should be this ensemble rather than just a single conformation. Because this ensemble of conformations is probably very narrowly distributed around the mean, it is often considered a safe assumption to ignore this source of complexity and concentrate on one representative conformation, which in all likelihood would approximate the mean of the ensemble.
Proteins participate in a second, much larger ensemble of conformations, usually referred to as the “denatured state” (2, 3, 4). In the past few years, considerable attention has been given to experimental and theoretical characterization of this complex and structurally diverse ensemble. Although current physical methods do not provide as high a resolution description of denatured states as they do for native states, the emerging picture is one of significant population of transient native-like local structures weakly coupled to each other (5, 6). An analysis of long range structure in an expanded denatured state of staphylococcal nuclease suggests that many of the global topological features of the native state are retained in the denatured state (7). In other words, the ensemble average structure of the denatured state resembles the native state, albeit at very low resolution.
In addition to forming a much more diverse ensemble, the conformations in the denatured state may have their structure and dynamic behavior determined by a smaller number of energy terms. Several authors have argued that burial of hydrophobic surface is the dominant force shaping structure in the denatured state ensemble (3, 5). In addition, the highly dynamic character and the much lower density of atoms suggest that dispersion forces, hydrogen bonds, and salt bridges may contribute little to the properties of denatured proteins. If the energetics are less dependent on the high resolution details of chain–chain interactions, the ensemble-averaged properties of the denatured state might be easier to predict than those of the native state. The database-derived energy/scoring functions currently used for structure prediction are thought to model primarily hydrophobic interactions (8, 9, 10) and thus may be suitable for prediction of structure in the denatured state ensemble.
If the ensemble-averaged topology (or low resolution structure) of the denatured state is approximately the same as that of the native state, the basin or minimum in the energy landscape containing both native and denatured states must have a partition function that is much larger than any other ensemble of structurally similar conformations. This idea is the fundamental hypothesis underlying the approach to structure prediction described in this paper.
RESULTS
Current knowledge of the residual structure in the denatured state and its energetic basis is too limited to reach definitive conclusions about the appropriateness of this large, dynamic ensemble as a target for predicting structural features of folded proteins. Therefore, we consider arguments concerning the structural correspondence between the denatured state and the native fold only as a general point of departure for the analysis reported here. In this spirit, we present two conjectures as working assumptions to be verified by future experimental and computational work rather than as established facts.
The first assumption is that the minimum in which native-like conformations reside is broader than any other minimum at the lowest energy levels in the folding landscape. The second assumption is that the breadth of this minimum results from the long range character of hydrophobic interactions and consequently should be detectable by using database-derived energy/scoring functions, which capture some of the features of hydrophobic interactions. Together, these two assumptions are equivalent to the statement that effective burial of hydrophobic residues can be retained throughout a larger range of structural perturbations of the native topology than of any other topology.
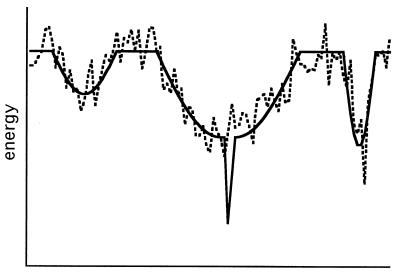
These assumptions are illustrated in a schematic energy landscape shown in Fig. 1, which positions the native state in a deep, narrow well located near the center of a broad, shallow minimum (solid line) (11). In a search to find the structure of a protein of known sequence, a relatively coarse grid search may generate multiple conformations within this broad minimum. Although an energy function that does not correctly quantify dispersion interactions, hydrogen bonds, and electrostatic interactions (Fig. 1, dashed line) may miss the steep drop in energy for conformations that comprise the native state, it may succeed in detecting the broad minimum if hydrophobic interactions are more or less correctly modeled.
Figure 1.
Schematic diagram of a hypothetical folding energy landscape. The x axis corresponds to a generalized structure coordinate (17, 26). The solid line corresponds to the internal free energy (17), and the dashed line corresponds to the value of a database-derived scoring function such as the one used in this work. The scoring function follows the true potential because it is sensitive to hydrophobic burial but produces noise and fails to detect the sharp drop in energy of the native state because of inaccuracies in quantifying hydrogen bonds, electrostatic, and van der Waals interactions. However, the scoring function is able to detect the higher density of low-energy states in the broad region surrounding the native state.
To the extent that these two assumptions are correct, protein structure at low resolution may be predicted by carrying out a coarse-grained sampling of conformational space and choosing the low-energy conformation having the largest number of structurally related low-energy conformations. In a situation such as that depicted in Fig. 1, relatively uniform sampling of conformation space followed by identification of the largest cluster of structurally related low-energy conformations would be expected to find the region of conformation space that contains the native state.
Two Sets of Computer-Generated “Decoy” Conformations for 10 Small Proteins.
To test this idea, we examined large sets of structures generated by Park and Levitt (12) for eight small proteins—cro repressor (2cro), a fragment of ribosomal protein L7/L12 (1ctf), the 434 repressor (1r69), calbindin (3icb), scorpion neurotoxin (1sn3), pancreatic trypsin inhibitor (4pti), ubiquitin (1ubq), and an electron transfer protein with an iron-sulfur center (4rxn). In brief, these structures were produced by an exhaustive search in which the angular relationships between five or six segments of fixed secondary structure were allowed to vary. After optimally fitting the native conformation as a trace of virtual Cα atoms with only four allowed torsion angles between residues, segments of the protein chain that closely followed a straight line (namely helices and strands) were identified, and residues between these straight segments became candidates for hinge angles (13). A total of 10 moveable residue positions were introduced as adjacent pairs in four or five hinge regions in each protein. Because torsion angles only were allowed to assume one of four possible values, each starting structure could be converted to 410 –1 alternative conformations by exhaustively enumerating all possible combinations of torsion angle values. After generating ≈1,000,000 conformations, the 80% with the greatest number of steric clashes were discarded, leaving a set of 200,000 decoy structures. After most remaining steric clashes were removed from these decoys by minimization in dihedral angle space, a more realistic chain representation was obtained by fitting backbone atoms (N, CA, C, O, CB) with correct bond distances and angles to the virtual Cα trace by using fragments of known proteins (K.T.S. and D.B., unpublished work).
A second, more structurally diverse set of conformations for four small all-helical proteins—staphylococcal protein A (1fc2), homeodomain repressor (1hdd), cro repressor (2cro), and calbindin (4icb)— also was analyzed. In previous work from this laboratory, ensembles of protein-like structures were generated by a Monte Carlo simulating annealing procedure in which segments of structure from the protein database were recombined to generate compact composites that scored well on the basis of a knowledge-based scoring function (13). To obtain local secondary structure compatible with the local structure, the protein segments used in this construction process were selected on the basis of similarity in amino acid sequence between the source protein and the target protein. To avoid biasing the generated set toward the wild-type conformation, all known structural homologues were removed from the set of proteins used as sources of structural segments. The 500 structures with the best overall score were saved for analysis. Unlike the conformations in the Park–Levitt sets, the conformations in the Simons sets showed considerable variability in the exact position of some helices (13).
The scoring functions used to evaluate the decoy structures were based on the decomposition
P(structure|sequence)≈P(sequence|structure)∗P(structure),
or Bayes’ rule, where P(x) is the a priori probability of the occurrence of x and P(y | x) is the conditional probability of y, given the occurrence of x. The first term on the right hand side quantifies the fit between the sequence and the structure and consisted of a residue-environment term that depends primarily on the hydrophobic interaction and a specific pair interaction term that captures interactions such as salt bridges and disulfide bonds. The second term on the right hand side is the probability that a candidate conformation is a properly folded protein structure. For scoring the Park–Levitt sets, P(structure) consisted of an excluded volume component plus a secondary structure packing term that is sensitive primarily to the relative orientation and packing of β strands (K.T.S. and D.B., unpublished work). For the Simons sets, this term only depended on excluded volume and the radius of gyration (13).
Results of Cluster Analysis.
Analysis of the eight Park–Levitt sets began with scoring each of the 200,000 conformations and saving the 1,000 with the best scores, which were defined as the low-energy ensemble for each protein sequence. The rms deviation (rmsd) of the Cα coordinates was calculated for each pair of conformations within a set, and the results stored in a 1,000 × 1,000 “distance matrix.” For each of a series of distance cutoffs ranging from 4 to 6 Å, the conformation having the most neighboring conformations within the distance cutoff was selected as the most central conformation. These results are listed in Table 1.
Table 1.
Clustering by structural similarity of the 1,000 lowest energy conformations in the Park–Levitt sets
| Protein | rmsd cutoff | Cluster size | rmsd center to native (rank in proximity to native state) | rmsd lowest energy conformation | Mean rmsd of ensemble |
|---|---|---|---|---|---|
| 2cro | 4 Å | 44 | 4.7 (44) | 5.6 | 8.8 |
| 5 Å | 85 | 3.2 | |||
| 6 Å | 151 | 6.7 | |||
| 1ctf | 4 Å | 69 | 1.7 (2) | 2.0 | 8.1 |
| 5 Å | 132 | 2.9 | |||
| 6 Å | 247 | 2.9 | |||
| 1r69 | 4 Å | 45 | 3.3 (12) | 5.2 | 8.0 |
| 5 Å | 129 | 4.2 | |||
| 6 Å | 257 | 3.9 | |||
| 3icb | 4 Å | 51 | 1.7 (1) | 4.7 | 9.2 |
| 5 Å | 83 | 2.0 | |||
| 6 Å | 137 | 1.7 | |||
| 1sn3 | 4 Å | 18 | 8.1 (417) | 2.1 | 8.4 |
| 5 Å | 40 | 7.2 | |||
| 6 Å | 120 | 6.9 | |||
| 4pti | 4 Å | 22 | 2.5 (10) | 10.0 | 9.2 |
| 5 Å | 44 | 5.0 | |||
| 6 Å | 100 | 6.1 | |||
| 1ubq | 4 Å | 52 | 2.0 (3) | 5.3 | 9.2 |
| 5 Å | 94 | 2.0 | |||
| 6 Å | 154 | 3.7 | |||
| 4rxn | 4 Å | 36 | 3.1 (13) | 8.4 | 8.4 |
| 5 Å | 82 | 3.2 | |||
| 6 Å | 153 | 5.4 |
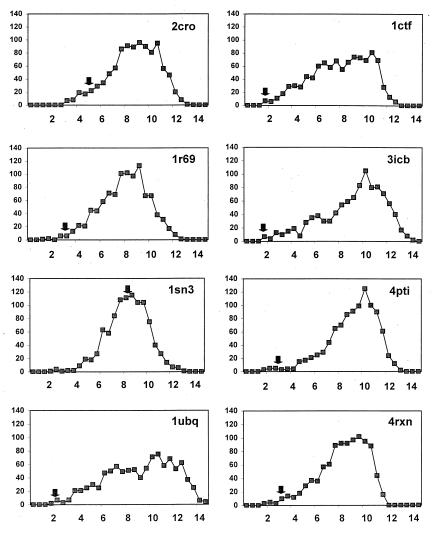
Fig. 2 shows the distribution of rmsd distances between the wild-type structure and each conformation in the set, along with the position of the center conformation for the largest cluster within 4-Å rmsd. As can be seen, in all cases but one (protein 1sn3, which has a very irregular structure held together by four disulfide bonds), the center conformation is significantly more similar to the native structure than the average member of the low-energy ensemble. In addition, for six of these seven cases, the center conformation was in the closest 1.5% of conformations with regard to rmsd from the native state.
Figure 2.
Histograms of the rmsd (Cα coordinates) from the native state to members of each of the Park–Levitt sets. An arrow marks the position of the center of the largest cluster of conformations by using a 4-Å rmsd cutoff. The bin intervals along the x axis are in 0.5-Å increments.
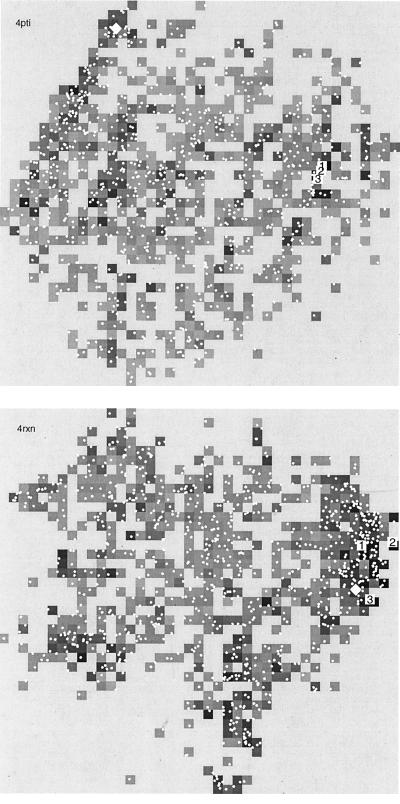
More graphic displays of the structural similarities among the 1,000 conformations in the Park–Levitt sets for proteins 4pti and 4rxn are shown in Fig. 3. By applying the statistical method of multidimensional scaling to the set of 500,000 pairwise distances, a map can be generated that represents an optimal solution of separating conformations in two dimensions in relationship to their distance in rmsd. The resulting physical distances between points on the map are not related linearly; only local rank ordering of distances is preserved by this scaling method. As can be seen, the conformations of very lowest energy are distributed fairly randomly over the maps of both proteins. Yet, in both cases, there is a significantly higher number density of low-energy conformations in a region near the native state.
Figure 3.
Multidimensional scaling maps of the ensemble of conformations in the Park–Levitt sets of conformations for 4pti (Upper) and 4rxn (Lower). The distance in rmsd between each pair of conformations is projected onto two dimensions, retaining relative distance relationships so that two structurally similar conformations tend to be located near each other. The position of each conformation is indicated by a small white dot. The position of the native state is marked with a white diamond, and the three conformations with three lowest (best) energy scores are marked with white boxes. The gray scale value of each pixel is determined by the lowest energy conformation within that small region of the map, with black being the very lowest energies.
To demonstrate that low energy plays an important role in these observed clusters of native-like conformations, the 200,000 conformations in each of the Park–Levitt sets were sorted by compactness, and the 1,000 most compact conformations were selected. Clustering this ensemble of conformations in the same manner gave the results seen in Table 2. For five of the proteins, the centers of the largest clusters are no more similar to the native structure than an average conformation within the ensemble.
Table 2.
Clustering by structural similarity of the 1,000 most compact conformations in the Park–Levitt sets
| Protein | rmsd cutoff | Cluster size | rmsd center to native | Mean rmsd of ensemble |
|---|---|---|---|---|
| 2cro | 4 Å | 68 | 3.0 | 8.2 |
| 5 Å | 123 | 2.3 | ||
| 6 Å | 210 | 4.1 | ||
| 1ctf | 4 Å | 33 | 8.6 | 9.6 |
| 5 Å | 43 | 11.4 | ||
| 6 Å | 84 | 7.2 | ||
| 1r69 | 4 Å | 18 | 9.7 | 8.8 |
| 5 Å | 31 | 3.7 | ||
| 6 Å | 87 | 2.9 | ||
| 3icb | 4 Å | 16 | 6.1 | 10.0 |
| 5 Å | 28 | 6.1 | ||
| 6 Å | 53 | 4.6 | ||
| 1sn3 | 4 Å | 18 | 9.0 | 9.4 |
| 5 Å | 61 | 9.0 | ||
| 6 Å | 149 | 8.1 | ||
| 4pti | 4 Å | 19 | 10.3 | 9.3 |
| 5 Å | 32 | 10.4 | ||
| 6 Å | 68 | 10.4 | ||
| 1ubq | 4 Å | 23 | 10.1 | 9.8 |
| 5 Å | 37 | 10.1 | ||
| 6 Å | 74 | 11.9 | ||
| 4rxn | 4 Å | 27 | 10.0 | 9.0 |
| 5 Å | 47 | 9.4 | ||
| 6 Å | 102 | 10.4 |
Surprisingly, for the three all-helical proteins, 2cro, 1r69, and 3icb, the cluster centers are considerably closer to the native structure than are the majority of configuration in the ensemble, although this trend may not be significant for 3icb. For 2cro, the centers of the 4-, 5-, and 6-Å groupings are closer to native than the cluster centers from the corresponding lowest energy ensemble. This may be a consequence of the fact that there are a limited number of ways to arrange helices to achieve a compact, self-avoiding configuration (15), and there are greater number of such well packed configurations around the native fold than other topological arrangements accessible in this ensemble.
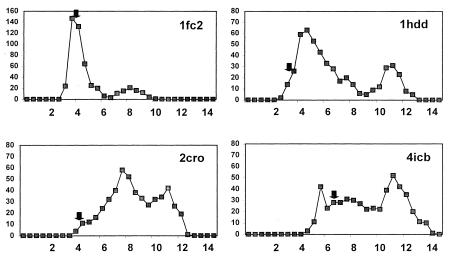
To analyze a second set of decoy structures constructed in an entirely different manner, the 500 conformations in the Simons sets also were clustered on the basis of structural similarity as measured by rmsd of the Cα coordinates (Table 3). As shown in Fig. 4, these four sets of proteins contained very few conformations within 3-Å rmsd of the native state. Nevertheless, for the two proteins 1hdd and 2cro, the center of the largest 4-Å cluster was among the top 5% in rmsd. For the 4icb set, which contained no member closer than 4-Å from the native state, the center of the largest 4-Å cluster had an rmsd from native of 6.2-Å, placing it only in the closest 20% of conformations. The fourth protein, 1fc2 or staphylococcal protein A, consists of a three-helical bundle. Not surprisingly, the level of structural diversity in the starting ensemble was relatively small. The bimodal distribution seen in Fig. 4 reflects the fact that there are only two topologies for packing three-helical bundles with very short connecting loops. The center of the largest cluster was only average in structural similarity to the native state, yet it did have the third helix on the correct side of the plane defined by the first two helices.
Table 3.
Clustering by structural similarity of the Simons sets of 500 conformations
| Protein | rmsd cutoff | Cluster size | rmsd center to native (rank in proximity to native state) | rmsd of lowest energy conformation | Mean rmsd of ensemble |
|---|---|---|---|---|---|
| 1fc2 | 4 Å | 410 | 4.0 (193) | 3.8 | 4.9 |
| 5 Å | 419 | 4.2 | |||
| 6 Å | 431 | 5.3 | |||
| 1hdd | 4 Å | 209 | 3.5 (17) | 5.2 | 6.8 |
| 5 Å | 296 | 4.5 | |||
| 6 Å | 348 | 4.8 | |||
| 2cro | 4 Å | 16 | 4.4 (3) | 7.9 | 8.7 |
| 5 Å | 37 | 7.2 | |||
| 6 Å | 90 | 4.9 | |||
| 4icb | 4 Å | 43 | 6.5 (82) | 5.8 | 9.4 |
| 5 Å | 89 | 7.0 | |||
| 6 Å | 143 | 5.9 |
Figure 4.
Histograms of the rmsd (Cα coordinates) from the native state to members of each of the Simons sets. An arrow marks the position of the center of the largest cluster of conformations by using a 4-Å rmsd cutoff. The bin intervals along the x axis are in 0.5-Å increments.
DISCUSSION
We describe a strategy for predicting protein structure at low resolution that goes beyond the standard approach of searching for the single lowest energy conformation. Instead of focusing on the lowest energy conformation, we search for the largest cluster of structurally related low-energy conformations. In all 12 sets of low-energy conformations studied, the conformation with the most other conformations within 4-Å rmsd was much more similar to the native structure than the majority of the conformations, and, in 9 of the 12 cases, this conformation was more similar to the native structure than the lowest energy conformation in the set.
Because the conformations in the Park–Levitt sets are rigidly fixed in secondary structure and have only four or five degrees of freedom for repositioning helices and strands, they correspond to a very limited search of conformation space. On the other hand, the algorithm used to generate the conformations in the Simons sets explores many more degrees of freedom. In this case, the type and position of secondary structures are constrained only by similarity in sequence between short segments of the target protein under construction and the template proteins from which structural segments were obtained. Thus, these sets represent a more realistic attempt to predict the structure of protein from sequence information alone. Overall, clustering of the Park–Levitt sets gave cluster centers closer to the native structure than did the Simons sets. Presumably, this is a consequence of the larger number of degrees of freedom used to generate the Simons sets. It will be important to determine in future work how readily the native minimum can be identified by using still more diverse conformational sampling strategies.
The higher density of low-energy conformations near the native structure is not an artifact built into these sets of conformations by the algorithms used to generate them. That the native state does not occupy a unique position in an ensemble is fairly obvious for the Simons sets. In this case, only the sequence of the target protein was used in the build-up process. Because the structural segments used in this process were derived from a subset of proteins that did not include known homologues, there should be no intrinsic bias toward over-representation of the tertiary structure of the native state among the conformations generated.
The conformations in the Park–Levitt sets, on the other hand, were derived from the native structure of the target protein, after it had been configured as a discreet state virtual Cα chain, by varying four or five hinge angles between fixed secondary structural segments. Because this construction process searched all allowed values of these angles, the resulting set of conformations is independent of the starting conformation. In other words, if one picked the lowest energy member of the ensemble and repeated the construction algorithm, exactly the same conformational set would be regenerated. Thus, there is no bias toward the more native-like members of the ensemble.
Why does clustering identify conformations considerably closer to the native structure than the conformation of lowest energy? One explanation is that the native topology provides the most robust arrangement of the chain for burying hydrophobic residues, in the sense that large structural perturbations can be tolerated without steric clashes and with relatively small increases in hydrophobic exposure. For example, in a four-helix bundle protein, relatively large translations of the helices relative to one another plus moderate rotations of the helices preserve hydrophobic burial. Similarly, in α/β sandwich proteins, the two layers may undergo rotations and translations relative to one another without exposing large amounts of hydrophobic surface. From the standpoint of the “new view” of protein folding (15–17), the greater breadth of the native minimum is a consequence of the assumption that native interactions are stronger on average than nonnative interactions, which results in a lowering of the energy of conformations with some native interactions formed. Our strategy also may be viewed as a type of signal averaging to compensate for noisy scoring functions, in which repeated independent attempts to find the native state are combined by picking the most common topology (the mode) rather than the lowest energy conformation.
The structural elements in native structures would be robust to displacement if they (i) often have sufficient local interactions to be low in energy in isolation; (ii) minimally restrict the ability of the remainder of the chain to form structural elements low in energy; and (iii) readily combine with other low-energy elements to form conformations that are low in energy. These features are consistent with the known modularity of structure in partially folded states of proteins—synthetic peptides, large protein fragments, and denatured proteins. Structural characterization of these types of systems have demonstrated that segments of a protein chain frequently have a high propensity in isolation to form local structures similar to those formed in the native protein (18–20).
Though limited to a very small sample, these results are encouraging and suggest that proteins in general may conform to some of the conditions we postulate might permit the prediction of structure at low resolution. If these results should prove to be general, they support the hypothesis that the native structures of proteins are in some sense surrounded by a large ensemble of low-energy conformations. In ascribing physical reality to this ensemble, we consider it most probable that it corresponds predominantly to the denatured state but also includes some high-energy forms of the native state involving large scale vibrational modes (21) plus partially unfolded states (22, 23).
Recently, the claim has been made that structures of naturally occurring proteins are selected by evolution because they have a high “designability,” i.e., a large tolerance to changes in amino acid sequence (a high sequence entropy). One plausible mechanism for such designability observed in simple lattice models is negative in character: minimization of the likelihood of favorable interactions in alternative structural states (24, 25). The results presented here suggest that a high tolerance of structural perturbation (high structural entropy) may be an additional, positive mechanism underlying tolerance of sequence perturbations.
Acknowledgments
The authors thank Ingo Ruczinski for the multidimensional scaling analysis shown in Fig. 3, Enoch Huang for helpful discussions and Britt Park and Michael Levitt for their decoy set. This work was supported by National Institutes of Health Grants GM34171 (to D.S.) and young investigator grants to D.B. from the National Science Foundation and the Packard Foundation. K.T.S. was supported by National Institutes of Health Training Grant PHS NRSA T32 GM07270.
ABBREVIATION
- rmsd
rms deviation
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Anfinsen C B. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Tanford C. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 3.Tanford C. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- 4.Shortle D. FASEB J. 1996;10:27–34. doi: 10.1096/fasebj.10.1.8566543. [DOI] [PubMed] [Google Scholar]
- 5.Dill K A, Shortle D. Annu Rev Biochem. 1991;60:795–825. doi: 10.1146/annurev.bi.60.070191.004051. [DOI] [PubMed] [Google Scholar]
- 6.Shortle D. Curr Opin Struct Biol. 1996;6:24–30. doi: 10.1016/s0959-440x(96)80091-1. [DOI] [PubMed] [Google Scholar]
- 7.Gillespie J, Shortle D. J Mol Biol. 1997;268:170–184. doi: 10.1006/jmbi.1997.0953. [DOI] [PubMed] [Google Scholar]
- 8.Thomas P D, Dill K A. J Mol Biol. 1996;257:457–469. doi: 10.1006/jmbi.1996.0175. [DOI] [PubMed] [Google Scholar]
- 9.Jernigan R L, Bahar I. Curr Opin Struct Biol. 1996;6:195–209. doi: 10.1016/s0959-440x(96)80075-3. [DOI] [PubMed] [Google Scholar]
- 10.Vajda S, Sippl M, Novotny J. Curr Opin Struct Biol. 1997;7:222–229. doi: 10.1016/s0959-440x(97)80029-2. [DOI] [PubMed] [Google Scholar]
- 11.Onuchic J N. Proc Natl Acad Sci USA. 1997;94:7129–7131. doi: 10.1073/pnas.94.14.7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park B, Levitt M. J Mol Biol. 1996;258:367–392. doi: 10.1006/jmbi.1996.0256. [DOI] [PubMed] [Google Scholar]
- 13.Simons K T, Kooperberg C, Huang E, Baker D. J Mol Biol. 1997;268:209–255. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 14.Murzin A G, Finkelstein A V. J Mol Biol. 1988;204:749–769. doi: 10.1016/0022-2836(88)90366-x. [DOI] [PubMed] [Google Scholar]
- 15.Bryngelson J D, Wolynes P G. Proc Natl Acad Sci USA. 1987;84:7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leopold P E, Montal M, Onuchic J N. Proc Natl Acad Sci USA. 1992;89:8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan H S, Dill K A. Prot Struct Funct Genet. 1998;32:2–33. doi: 10.1002/(sici)1097-0134(19980101)30:1<2::aid-prot2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 18.Dobson C M. Curr Opin Struct Biol. 1992;2:6–12. [Google Scholar]
- 19.Shortle D, Wang Yi, Gillespie J, Wrabl J O. Protein Sci. 1996;5:991–1000. doi: 10.1002/pro.5560050602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulman B A, Kim P S, Dobson C M, Redfield C. Nat Struct Biol. 1997;4:630–634. doi: 10.1038/nsb0897-630. [DOI] [PubMed] [Google Scholar]
- 21.Tolman J R, Flanagan J M, Kennedy M A, Prestegard J H. Nat Struct Biol. 1997;4:292–297. doi: 10.1038/nsb0497-292. [DOI] [PubMed] [Google Scholar]
- 22.Bai Y, Sosnick T R, Mayne L, Englander S W. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chamberlain A K, Handel T M, Marqusee S. Nat Struct Biol. 1996;3:782–787. doi: 10.1038/nsb0996-782. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Helling R, Tang C, Wingreen N. Science. 1996;273:666–669. doi: 10.1126/science.273.5275.666. [DOI] [PubMed] [Google Scholar]
- 25.Yue K, Dill K A. Proc Natl Acad Sci USA. 1995;92:146–150. doi: 10.1073/pnas.92.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bryngelson J D, Onuchic J N, Socci N D, Wolynes P G. Protein Struct Funct Genet. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]