Abstract
The specificities of 65 compounds reported to be relatively specific inhibitors of protein kinases have been profiled against a panel of 70–80 protein kinases. On the basis of this information, the effects of compounds that we have studied in cells and other data in the literature, we recommend the use of the following small-molecule inhibitors: SB 203580/SB202190 and BIRB 0796 to be used in parallel to assess the physiological roles of p38 MAPK (mitogen-activated protein kinase) isoforms, PI-103 and wortmannin to be used in parallel to inhibit phosphatidylinositol (phosphoinositide) 3-kinases, PP1 or PP2 to be used in parallel with Src-I1 (Src inhibitor-1) to inhibit Src family members; PD 184352 or PD 0325901 to inhibit MKK1 (MAPK kinase-1) or MKK1 plus MKK5, Akt-I-1/2 to inhibit the activation of PKB (protein kinase B/Akt), rapamycin to inhibit TORC1 [mTOR (mammalian target of rapamycin)–raptor (regulatory associated protein of mTOR) complex], CT 99021 to inhibit GSK3 (glycogen synthase kinase 3), BI-D1870 and SL0101 or FMK (fluoromethylketone) to be used in parallel to inhibit RSK (ribosomal S6 kinase), D4476 to inhibit CK1 (casein kinase 1), VX680 to inhibit Aurora kinases, and roscovitine as a pan-CDK (cyclin-dependent kinase) inhibitor. We have also identified harmine as a potent and specific inhibitor of DYRK1A (dual-specificity tyrosine-phosphorylated and -regulated kinase 1A) in vitro. The results have further emphasized the need for considerable caution in using small-molecule inhibitors of protein kinases to assess the physiological roles of these enzymes. Despite being used widely, many of the compounds that we analysed were too non-specific for useful conclusions to be made, other than to exclude the involvement of particular protein kinases in cellular processes.
Keywords: anti-cancer drugs, drug discovery, inhibitor specificity, kinase profiling, protein kinase
Abbreviations: AICAR, aminoimidazole-4-carboxamide-1-β-D-ribofuranoside; ATF2, activating transcription factor 2; ATM, ataxia telangiectasia mutated; EGF, epidermal growth factor; AMPK, AMP-activated protein kinase; BRSK, brain-specific kinase; CAK, cyclin-dependent kinase-activating kinase; CaMK, calmodulin-dependent kinase; CaMKK, CaMK kinase; CDK, cyclin-dependent protein kinase; CHK, checkpoint kinase; CK, casein kinase; CSK, C-terminal Src kinase; DYRK, dual-specificity tyrosine-phosphorylated and -regulated kinase; EF2K, elongation-factor-2 kinase; Eph-A2, Ephrin A2 receptor; ERK, extracellular-signal-regulated kinase; FGF-R, fibroblast-growth-factor receptor; FKBP, FK506-binding protein; FMK, fluoromethylketone; GAK, cyclin G-associated kinase; GSK3, glycogen synthase kinase 3; GST, glutathione transferase; HEK-293 cells, human embryonic kidney-293 cells; HIPK, homeodomain-interacting protein kinase; His6, hexahistidine; IGF-1, insulin-like growth factor-1; IKK, inhibitory κB kinase; IL-1, interleukin 1; JNK, c-Jun N-terminal kinase; Lck, lymphocyte cell-specific protein-tyrosine kinase; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; MAPKAP-K, MAPK-activated protein kinase; MARK, microtubule-affinity-regulating kinase; MBP, myelin basic protein; MELK, maternal embryonic leucine-zipper kinase; MKK1, MAPK kinase-1 (also called MEK1, MAPK or ERK kinase 1); MMS, methyl methanesulfonate; MNK, MAPK-integrating protein kinase; MSK, mitogen- and stress-activated protein kinase; MST, mammalian homologue Ste20-like kinase; NDRG, N-myc downstream-regulated gene; NEK, NIMA (never in mitosis in Aspergillus nidulans)-related kinase; NFAT, nuclear factor for activated T-cells; PAK, p21-activated protein kinase; PDK, 3-phosphoinositide-dependent protein kinase; PH, pleckstrin homology; PHK, phosphorylase kinase; PI3K, phosphatidylinositol (phosphoinositide) 3-kinase; PIM, provirus integration site for Moloney murine leukaemia virus; PKA, cAMP-dependent protein kinase; PKB, protein kinase B (also called Akt); PKC, protein kinase C; PKD, protein kinase D; PLK, polo-like kinase; PPAR, peroxisome-proliferator-activated receptor; PRAK, p38-regulated activated kinase; PRK, protein kinase C-related kinase; PTEN, phosphatase and tensin homologue deleted on chromosome 10; RIP2, receptor-interacting protein 2; ROCK, Rho-dependent protein kinase; RSK, p90 ribosomal S6 kinase; S6K1, S6 kinase 1; Sf21, Spodoptera frugiperda (fall armyworm) 21; SGK, serum- and glucocorticoid-induced kinase; SmMLCK, smooth-muscle myosin light-chain kinase; Src, sarcoma kinase; Src-I1, Src inhibitor 1; SRPK, serine-arginine protein kinase; TANK, TRAF (tumour-necrosis-factor-receptor-associated factor)-family-member-associated nuclear factor κB activator; TBK1, TANK-binding kinase 1; TORC1, mTOR (mammalian target of rapamycin)–raptor (regulatory associated protein of mTOR) complex; VEGF, vascular endothelial growth factor (vasoendothelial growth factor); Yes1, Yamaguchi sarcoma viral oncogene homologue 1; ZMP, aminoimidazole-4-carboxamide-1-β-D-ribofuranoside monophosphate
INTRODUCTION
Small cell-permeant inhibitors of protein kinases have become invaluable reagents with which to investigate the physiological roles of protein kinases, because they can be used simply and rapidly to block endogenous kinase activity in normal cells and tissues, as well as transformed cell lines. In recent years a plethora of protein kinase inhibitors have become available commercially, and researchers are often faced with a bewildering variety of compounds from which to choose from, each compound being purported to be a ‘specific’ inhibitor of a particular protein kinase. It is therefore difficult to decide which compound will switch off the activity of the protein kinase or signalling pathway under investigation, both effectively and specifically.
There are some 500 protein kinases encoded by the human genome, most of which are members of the same superfamily, so that the issue of selectivity is critical. Seven years ago we studied 28 commonly used protein kinase inhibitors and examined their specificities against a panel of 24 different protein kinases [1], and a few years later we extended this analysis to a further 14 compounds against a slightly larger panel [2]. These studies revealed that a number of ‘specific’ inhibitors affected so many protein kinases as to render meaningless the conclusions made about the role of a particular kinase by the use of these compounds. These studies appear to have been useful to the cell-signalling community, as judged by the number of times that the first paper [1] was downloaded from the Biochemical Journal website in 2004 (7600 times) and cited in other papers (over 1500 times).
Over the past few years, we have increased the size of our core ‘profiling’ panel from 30 to over 70 protein kinases and have used this enlarged panel to examine further the specificities of many protein kinase inhibitors. Here we present information about the specificities of 65 inhibitors and make recommendations about their use. It should be noted that each protein kinase was assayed at or below the Km for ATP, explaining why the IC50 values for some protein kinase inhibitors are lower than those reported in previously published papers where a higher (0.1 mM) concentration of ATP was employed in the assays. These lower concentrations of ATP not only allow a more stringent test of the specificities of protein kinase inhibitors, but also reduce the cost of performing this expensive analysis.
MATERIALS AND METHODS
Protein kinase inhibitors and other materials
SB 203580, SB 202190, PP1, PP2, NA-PP1, NM-PP1, SU 6656, Src inhibitor-1, ZM 336372, alsterpaullone, kenpaullone, LY 294002, Akt-I-1,2, rapamycin, IC 261, roscovitine, purvalanol, PS 1145, STO 609, SC 514, SP 600125, AS 601245, UCN01, Ro 318220, Go 6976, KT 5720, Rottlerin, H7, H8, H89, HA 1077, H 1152, Y27632 and Compound C were purchased from Calbiochem, GW 5074, SB 216763, SB 415286 and wortmannin were from Sigma, harmine, harmalol, harmane and harmaline were from Fluka, U0126 was from Promega, and CK1-7 was from Seikegaku Corp, Tokyo, Japan. SL0101 was purchased from Toronto Research Chemicals, and one sample was a gift from Dr Morten Frodin, Biotech Research and Innovation Center, Copenhagen Biocenter, Copenhagen, Denmark. LY333531 was a gift from Dr Alex Kozikowski (College of Pharmacy, University of Chicago at Illinois, Chicago, IL, U.S.A.), BAY 439006 was a gift from Dr Richard Marais (Institute for Cancer Research, London, U.K.), and FMK (fluoromethylketone) was a gift from Dr Jack Taunton (Department of Cellular and Molecular Pharmacology, University of California San Francisco, San Francisco, CA, U.S.A.).
BIRB 0796 [3], PD 184352 [4], PD 0325901 and PD 0325901-Cl [5], CT 99021[6], BI D1870 [7], AR-A0-14418 [8], PI 103 [9,10], A-443654[11–13], D4476 [14–16], VX680 [17], BMS-345541 [18], CGP 57380 [19], BX 795 and BX 320 [20], and SU6668 [21] were synthesized using the methods indicated. The structures of the compounds that were synthesized are shown in Supplementary Figure S1 at http://www.BiochemJ.org/bj/408/bj408ppppadd.htm.
MMS (methyl methanesulfonate) was from Sigma, IGF-1 (insulin-like growth factor 1) and EGF (epidermal growth factor) were from Invitrogen, an antibody that recognizes the phosphorylated and unphosphorylated forms of ERK5 (extracellular-signal-related kinase 5) equally well and phospho-specific antibodies that recognize CHK1 (checkpoint kinase 1) phosphorylated at Ser345, CHK2 at Thr68, PKB (protein kinase B/Akt) at Ser473, and the phosphorylated forms of ERK1 and ERK2, were from Cell Signaling Technologies.
Source and purification of kinases
Unless stated otherwise, all protein kinases were of human origin and encoded full-length proteins. Apart from the AMPK (AMP-activated protein kinase) complex, which was purified from rat liver, all other proteins were either expressed as GST (glutathione transferase) fusion proteins in Escherichia coli or as hexahistidine (His6)-tagged proteins in Sf21 (Spodoptera frugiperda 21) insect cells. GST fusion proteins were purified by affinity chromatography on glutathione–Sepharose, and His6-tagged proteins on nickel/nitrilotriacetate–agarose. The procedures for expressing some of the protein kinases used in the present study have been detailed previously [1,2]. GAK (cyclin G-associated kinase) expressed in E. coli was a gift from Marjan Ford, MRC Laboratory of Molecular Biology, Cambridge, U.K., whereas IKKα [IκB (inhibitory κB) kinase] was purchased from Upstate (now part of Millipore). The following sections outline the DNA vectors synthesized and the procedures used to express and purify protein kinases that have not been reported previously.
Expression of recombinant proteins in E. coli
The following protein kinases were expressed in E. coli: CHK2[5–543], CK1δ[1–294] (casein kinase[1–294]), cyclin A2[171–432], CDK2 (cyclin-dependent protein kinase 2), CAK (CDK-activating kinase; also called CDK7) with an additional His6 tag at its C-terminus, PKA (cAMP-dependent protein kinase), PHK[2–297] (phosphorylase kinase[2–297]), CaMK-1 (calmodulin-dependent kinase 1), EF2K (elongation factor 2 kinase), JNK3α1[40–422] (c-jun N-terminal kinase 3[40–422]), the JNK1[M108A] and JNK1[M108G] mutants, MAPKAP-K2[46–400] {MAPK (mitogen-activated protein kinase)-activated protein kinase-2[46–400]} and MAPKAP-K3, smMLCK[475–838] (smooth-muscle myosin light-chain kinase[475–838]), MNK1 and MNK2 (MAPK-interacting kinases 1 and 2), PIM2 (provirus integration site for Moloney murine leukaemia virus 2), SRPK1 (serine-arginine protein kinase 1), DYRK1A[1–499] (dual-specificity tyrosine-phosphorylated and -regulated kinase 1[1–499]), DYRK2 and DYRK3, PAK4, PAK5 and PAK6 (p21-activated kinases 4, 5 and 6), CaMKKα and CaMKKβ (calmodulin-dependent kinase kinases α and β), MELK (maternal embryonic leucine-zipper kinase), ERK1 (extracellular-signal-regulated kinase 1) and HIPK2[165–564] (homeodomain-interacting protein kinase 2[165–564]) and HIPK3[161–562].
Expression of recombinant proteins in Sf21 cells
The following protein kinases were expressed in insect Sf21 cells: RSK1 (p90 ribosomal S6 kinase-1), RSK2, NEK2a [NIMA (never in mitosis in Aspergillus nidulans)-related protein kinase 2a], NEK6[8–313] and NEK7, PKCα (protein kinase Cα), Aurora B and Aurora C, ERK8, IKKβ, MARK3 (microtubule-affinity-regulating kinase 3), MST2 (mammalian Ste20-like kinase 2), PKBα[118–480][S473D], PKBβ[120–481][S474D], PDK1[52–556] (3-phosphoinositide-dependent protein kinase-1[52–556]), PKD1 (protein kinase D1; also known as PKCμ), PLK1 (polo-like kinase 1), PRK2[501–984] (PKC-related kinase 2[501–984]), ROCK2[2–543] (Rho-dependent protein kinase 2[2–543]), SGK1[60–431][S422D] (serum- and glucocorticoid-induced kinase-1[60–431][S422D]), S6K1[1–421][T412E] (S6 kinase 1[1–421][T412E]), Src (chicken), JNK2α2 (c-Jun N-terminal kinase 2), PIM1, PIM3, BRSK2 (brain-specific kinase 2), PKCζ, mouse Lck (lymphocyte cell-specific protein-tyrosine kinase), c-Raf[306–648][Y340D/Y341D/V492E] (these mutations produce a constitutively active enzyme of high specific activity), B-Raf[2–766][V600E] (the activated oncogenic mutant found in many malignant melanomas), RIP2[2–540] (receptor-interacting protein 2[2–540]; also called RICK and CARDIAK), IKKϵ, TBK1 (TANK-binding kinase 1), Yes (Yamaguchi sarcoma viral oncogene homologue 1), FGFR1[400–820] (fibroblast-growth-factor receptor 1[400–820]) and Ephrin A2[591–976].
Activation of protein kinases
In order to produce activated forms of Aurora B and Aurora C, insect Sf21 cells were incubated for 1 h with the protein phosphatase inhibitor okadaic acid (50 nM), whereas, to produce activated PLK1, the Sf21 cells were incubated for 4 h with 100 nM okadaic acid prior to harvesting the cells and purifying the enzyme. MKK1 (MAPK kinase-1) was activated with c-Raf, wild-type and mutant JNK isoforms with MKK4 and MKK7, p38 MAPK isoforms with MKK6, MAPKAP-K2, MAPKAP-K3, PRAK (p38-regulated activated kinase), MNK1, MNK2 and MSK1 with p38α MAP kinase, RSK1 and RSK2 with ERK2 plus PDK1; PKBα, PKBβ, SGK1 and S6K1 with PDK1, and ERK1 and ERK2 with MKK1. To activate CDK2, bacterial pellets expressing cyclin A2 and CDK2 were mixed together, lysed, then purified on glutathione–Sepharose. The GST tags were removed by cleavage with PreScission protease and the CDK2–cyclin A2 complex was purified by chromatography on SP (sulfopropyl)-Sepharose. It was then activated with CAK1/CDK7 followed by chromatography on nickel-nitrilotriacetate–agarose to remove CAK1/CDK7, which binds to this column by virtue of its C-terminal His6 tag. All the other protein kinases were active as expressed.
Protein kinase assays
All assays (25.5 μl volume) were carried out robotically at room temperature (21 °C) and were linear with respect to time and enzyme concentration under the conditions used. Assays were performed for 30 min using Multidrop Micro reagent dispensers (Thermo Electron Corporation, Waltham, MA, U.S.A.) in a 96-well format. The concentration of magnesium acetate in the assays was 10 mM and [γ-33P]ATP (800 c.p.m./pmol) was used at 5, 20 or 50 μM as indicated, in order to be at or below the Km for ATP for each enzyme. Protein kinases assayed at 5 μM ATP were: MKK1, ERK1, p38γ MAPK, p38δ MAPK, ERK8, PKBα, PKCζ, PRK2, GSK3β, CK2, MARK3, IKKβ, DYRK3, PIM2, EF2K, PLK1, Aurora C, HIPK2 and PAK4. Protein kinases assayed at 20 μM ATP were: JNK1, JNK2, p38β MAPK, PDK1, SGK1, S6K1, PKA, ROCK2, PKCα, MSK1, MAPKAP-K2, MAPKAP-K3, PRAK, CaMKKα, CaMKKβ, CHK1, CHK2, CDK2, Aurora B, CK1, PIM1, PIM3, NEK7, MST2, HIPK3, PAK5, PAK6, CSK, Yes and FGF-R1. Protein kinases assayed at 50 μM ATP were: Eph-A2 (Ephrin-A2 receptor), ERK2, JNK3, p38α MAPK, RSK1, RSK2, PKBβ, PKD1, MNK1, MNK2, AMPK, CaMK1, smMLCK, PHK, BRSK2, MELK, DYRK1a, DYRK2, NEK2a, NEK6, SRPK1, Src, Lck, IKKϵ and TBK1. Protein kinases assayed at 0.1 mM ATP were RIP2, GAK, c-Raf and B-Raf.
The assays were initiated with MgATP, stopped by the addition of 5 μl of 0.5 M orthophosphoric acid and spotted on to P81 filter plates using a unifilter harvester (PerkinElmer, Boston, MA, U.S.A.). The IC50 values of inhibitors were determined after carrying out assays at ten different concentrations of each compound.
PKA was assayed against the substrate peptide LRRASLG (300 μM), PKCα and GAK against the protein histone H1 (0.1 mg/ml for PKCα and 1.0 mg/ml for GAK), PHK against the substrate peptide KRKQISVRGL (300 μM), NEK2a against the peptide RFRRSRRMI (300 μM), NEK6 and NEK7 against the peptide FLAKSFGSPNRAYKK (300μM), ROCK and PRK2 against a peptide corresponding to the C-terminal region of ribosomal protein S6 (KEAKEKRQEQIAKRRRLSSLRASTSKSGGSQK) (30 μM). Aurora B and Aurora C were both assayed against the substrate peptide LRRLSLGLRRLSLGLRRLSLGLRRLSLG (300 μM), ERK1, ERK8, HIPK1, HIPK3, MST-2, IKKα and IKKϵ against MBP (myelin basic protein; 0.33 mg/ml), RIP2 against MBP (1.0 mg/ml), IKKβ against the peptide LDDRHDSGLDSMKDEEY (300 μM), and JNK2 and JNK3 against ATF2[19–96] (activating transcription factor 2[19–96]; 3 μM). MARK3 was assayed against the peptide KKKVSRSGLYRSPSMPENLNRPR (300 μM), RSK1, RSK2, MAPKAP-K3 and PKD1 against KKLNRTLSVA (30 μM), MNK1 and MNK2 against the eIF4E (eukaryotic translation initiation factor 4E) protein (0.5 mg/ml), EF2K assayed against the peptide RKKFGESKTKTKEFL (300 μM) and PIM1, PIM2 and PIM3 against RSRHSSYPAGT (300 μM). PKBβ was assayed against the peptide GRPRTSSFAEGKK (30 μM), PLK1 against ISDELMDATFADQEAKKK (300 μM), Src against KVEKIGEGTYGVVYK (300 μM), CaMK-1 against YLRRRLSDSNF (300 μM), smMLCK against KKRPQRATSNVFA (300 μM) and SRPK1 against RSRSRSRSRSRSRSR (300 μM). DYRK1A, DYRK2 and DYRK3 were both assayed against Woodtide (KKISGRLSPIMTEQ) (300 μM), whereas PAK4, 5 and 6 were assayed against RRRLSFAEPG (300 μM). CaMKKα, CaMKKβ and TBK1 were assayed against AKPKGNKDYHLQTCCGSLAYRRR (300 μM), MELK and BRSK2 against KKLNRTLSFAEPG (300 μM) and PKCζ against ERMRPRKRQGSVRRV (300 μM). The protein tyrosine kinases Yes, FGF-R1 and Ephrin A2 were assayed with poly(Glu4-Tyr1) (1 mg/ml). The substrates used for other protein kinases were described previously[1,2].
Unless stated otherwise, enzymes were diluted in a buffer consisting of 50 mM Tris/HCl, pH 7.5, 0.1 mM EGTA, 1 mg/ml BSA and 0.1% 2-mercaptoethanol and assayed in a buffer comprising 50 mM Tris/HCl, pH 7.5, 0.1 mM EGTA and 0.1% 2-mercaptoethanol. For CaMK1 and CaMKK isoforms, the assay mixtures also contained 0.5 mM CaCl2 and 0.3 μM calmodulin. PKCα was diluted into 20 mM Hepes (pH 7.4)/0.03 Triton X-100 and assayed in the same buffer containing 0.1 mg/ml phosphatidylserine, 10 μg/ml diacylglycerol and 0.1 mM CaCl2. PHK (5–20 m-units) was diluted in 50 mM sodium β-glycerophosphate (pH 7.0)/0.1% 2-mercaptoethanol and assayed in a buffer comprising 50 mM Tris/HCl, 50 mM sodium β-glycerophosphate, pH 8.2, and 0.04 mM CaCl2. EF2K (5–20 m-units) was diluted into 50 mM Hepes (pH 6.6)/0.1% 2-mercaptoethanol/1.0 mg/ml BSA and assayed in the same buffer containing 0.2 mM CaCl2 and 0.3μM calmodulin. smMLCK (5–20 m-units) was diluted in 50 mM Hepes (pH 7.5)/0.1 mM EGTA/1.0 mg/ml BSA/0.1% 2-mercaptoethanol and assayed in the same buffer containing 5 mM CaCl2 and 10 μM calmodulin. PKA (5–20 m-units) was diluted in 20 mM Mops (pH 7.5)/1 mM EGTA/0.01% Brij 35/1.0 mg/ml BSA/0.1% 2-mercaptoethanol and assayed in 8 mM Mops (pH 7.5)/0.2 mM EDTA. The protein kinases c-Raf and B-Raf were assayed as described previously [22].
RESULTS AND DISCUSSION
Inhibitors of p38 MAPK (SB 203580, SB 202190, BIRB 0796)
SB 203580 [23] and its close relative SB 202190 have been exploited in thousands of reported studies to assess the physiological roles of p38α and p38β MAPKs. Although these compounds have been, and still are, very useful, more recent studies have identified other protein kinases that they inhibit with similar (GAK and CK1) or even greater (RIP2) potency [24]. SB 203580 also inhibits c-Raf [25] and GSK3 in vitro (Table 1), albeit less strongly, and inhibits the formation of ZMP (aminoimidazole-4-carboxamide-1-β-D-ribofuranoside monophosphate), an activator of AMPK, from its inactive precursor AICAR (aminoimidazole-4-carboxamide-1-β-D-ribofuranoside), probably by inhibiting adenosine transporters [26]. Thus there is a danger that the observed effects of SB 203580/SB 202190 on cells result from the inhibition of a target(s) distinct from p38α/p38β MAPKs. This inherent problem can be overcome by examining whether the effects of these compounds are no longer observed in cells that express an SB 203580-resistant mutant of p38α MAPK [27] or p38β MAPK, or by studying whether the results obtained with SB 203580 are also observed in cells from knockout mice that do not express p38α MAPK (see, e.g., [28]) and/or p38β MAPK. However, although p38β MAPK-deficient mice are viable, p38α MAPK-deficient mice display embryonic lethality, and studies with p38α MAPK knockout cells have so far been confined to the use of embryonic fibroblasts. The availability of inhibitors that are more specific than SB 203580 and SB 202190 would therefore be very useful.
Table 1. Specificities of compounds developed as inhibitors of p38α MAPK, Raf and Src family kinases.
The concentrations of compounds used in the assays are indicated below each substrate name, and the results shown are percentage activity (averages of duplicate determinations) remaining in the presence of inhibitor as compared with the control incubations with the inhibitor omitted. Each experiment was repeated two or three times with similar results. Further details of the assays are given in the Materials and methods section. Important values are highlighted in boldface type. Abbreviations: ND, not determined.
| Percentage activity remaining | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kinase | SB 203580 (1 μM) | SB 202190 (1 μM) | BIRB 0796 (1 μM) | SU 6656 (1 μM) | Src-I1 (1 μM) | PP1 (1 μM) | PP2 (1 μM) | NA-PP1 (1 μM) | NA-PP1 (10 μM) | NM-PP1 (1 μM) | NM-PP1 (10 μM) | ZM 336372 (10 μM) | BAY 439006 (10 μM) | GW 5074 (1 μM) |
| MKK1 | 85±8 | 70±9 | 55±4 | 78±2 | 50±8 | 67±5 | 76±2 | 86±5 | 65±4 | 113±2 | 76±7 | 106±1 | 74±3 | 73±7 |
| ERK1 | 89±0 | 104±0 | 97±0 | 115±1 | 103±8 | 113±1 | 104±1 | 104±1 | 70±1 | 85±6 | 46±2 | 81±1 | 92±7 | 99±3 |
| ERK2 | 83±7 | 90±8 | 95±10 | 95±1 | 75±9 | 94±2 | 85±1 | 82±5 | 62±1 | 65±4 | 32±8 | 93±3 | 91±3 | 89±6 |
| JNK1 | 95±0 | 83±2 | 90±2 | 94±5 | 84±3 | 93±10 | 92±4 | 92±2 | 93±2 | 90±0 | 85±4 | 85±2 | 83±5 | 94±5 |
| JNK2 | 60±3 | 42±1 | 4±0 | 87±1 | 94±9 | 85±4 | 81±5 | 85±1 | 57±4 | 79±1 | 64±5 | 85±6 | 65±4 | 96±8 |
| JNK3 | 72±7 | 49±0 | 69±0 | 95±0 | 81±7 | 85±0 | 66±0 | 93±3 | 88±3 | 98±9 | 80±1 | 86±0 | 101±4 | 91±2 |
| p38α MAPK | 9±0 | 3±0 | 4±0 | 85±2 | 83±4 | 43±2 | 49±4 | 49±2 | 17±5 | 41±2 | 8±2 | 8±1 | 14±2 | 77±2 |
| P38β MAPK | 13±1 | 5±3 | 13±4 | 87±5 | 90±8 | 38±2 | 48±0 | 77±2 | 24±2 | 53±5 | 15±5 | 15±2 | 22±4 | 100±4 |
| p38γ MAPK | 104±1 | 86±7 | 36±4 | 102±3 | 104±2 | 98±8 | 107±2 | 97±1 | 81±2 | 96±1 | 99±2 | 95±1 | 68±1 | 107±7 |
| p38δ MAPK | 107±3 | 87±1 | 40±5 | 86±1 | 86±2 | 69±7 | 93±2 | 108±1 | 80±2 | 98±1 | 94±4 | 95±9 | 61±4 | 97±4 |
| ERK8 | 93±9 | 106±3 | 95±6 | 66±1 | 82±7 | 94±2 | 107±3 | 81±1 | 59±5 | 77±0 | 49±0 | 88±1 | 7±2 | 68±2 |
| RSK1 | 52±8 | 78±2 | 35±2 | 56±1 | 73±5 | 94±4 | 118±0 | 89±3 | 53±5 | 84±2 | 56±7 | 70±1 | 41±4 | 65±2 |
| RSK2 | 93±6 | 89±1 | 49±7 | 50±5 | 80±2 | 99±1 | 86±6 | 96±2 | 71±1 | 98±1 | 68±2 | 69±4 | 54±3 | 70±2 |
| PDK1 | 81±2 | 93±0 | 86±6 | 97±9 | 88±3 | 84±11 | 88±6 | 98±5 | 98±3 | 97±1 | 102±0 | 84±3 | 82±2 | 98±4 |
| PKBα | 89±11 | 89±3 | 90±8 | 95±5 | 80±12 | 95±0 | 98±0 | 102±9 | 50±1 | 83±2 | 63±2 | 105±1 | 94±1 | 90±3 |
| PKBβ | 90±1 | 91±6 | 89±2 | 86±4 | 86±7 | 97±11 | 90±1 | 96±2 | 89±1 | 98±5 | 92±4 | 63±1 | 83±1 | 82±5 |
| SGK1 | 91±0 | 76±4 | 72±2 | 98±8 | 106±4 | 87±0 | 88±6 | 102±5 | 92±2 | 99±2 | 100±5 | 91±2 | 90±3 | 82±1 |
| S6K1 | 80±5 | 83±1 | 99±6 | 83±1 | 92±4 | 42±5 | 61±5 | 86±7 | 61±7 | 91±7 | 82±7 | 111±3 | 76±4 | 98±6 |
| PKA | 89±4 | 73±1 | 69±4 | 98±6 | 95±9 | 52±3 | 50±5 | 86±3 | 56±2 | 32±2 | 7±1 | 93±6 | 72±3 | 109±2 |
| ROCK 2 | 73±13 | 84±4 | 79±2 | 64±0 | 84±10 | 81±0 | 90±0 | 86±2 | 30±1 | 84±1 | 77±1 | 96±3 | 71±8 | 66±1 |
| PRK2 | 95±8 | 65±4 | 74±7 | 37±7 | 81±4 | 84±3 | 104±7 | 110±1 | 90±2 | 109±5 | 98±2 | 92±6 | 79±1 | 91±6 |
| PKCα | 86±6 | 69±1 | 62±3 | 92±1 | 95±2 | 75±4 | 71±3 | 78±4 | 40±9 | 82±1 | 80±2 | 89±5 | 87±1 | 92±7 |
| PKCζ | 89±0 | 113±5 | 116±6 | 97±3 | 95±4 | 93±7 | 86±6 | 86±0 | 84±1 | 83±2 | 85±5 | 78±8 | 92±4 | 103±9 |
| PKD1 | 78±7 | 61±4 | 93±7 | 90±1 | 81±4 | 76±2 | 75±8 | 35±2 | 8±3 | 13±0 | 3±1 | 104±3 | 90±1 | 99±4 |
| MSK1 | 91±10 | 87±0 | 84±5 | 88±1 | 81±3 | 72±12 | 83±1 | 81±6 | 47±6 | 78±1 | 70±3 | 96±1 | 84±3 | 80±4 |
| MNK1 | 109±10 | 87±5 | 79±2 | 88±2 | 81±1 | 76±1 | 78±7 | 110±1 | 96±1 | 110±9 | 106±0 | 86±1 | 67±5 | 90±6 |
| MNK2 | 107±1 | 103±6 | 91±0 | 102±2 | 61±7 | 97±1 | 107±1 | 106±1 | 85±8 | 115±1 | 98±5 | 102±8 | 40±3 | 91±5 |
| MAPKAP-K2 | 84±2 | 122±4 | 105±3 | 107±7 | 81±3 | 101±1 | 72±1 | 117±0 | 98±4 | 99±3 | 115±7 | 89±6 | 104±5 | 84±7 |
| MAPKAP-K3 | 120±5 | 104±2 | 89±3 | 105±9 | 88±4 | 98±3 | 101±1 | 91±1 | 88±5 | 96±1 | 94±2 | 105±8 | 92±2 | 95±5 |
| PRAK | 96±5 | 84±0 | 94±5 | 68±6 | 94±5 | 101±0 | 92±8 | 89±8 | 76±0 | 90±1 | 62±1 | 61±7 | 84±5 | 97±8 |
| CaMKKα | 106±14 | 94±1 | 96±4 | 20±4 | ND | 92±17 | 99±1 | 94±6 | 76±9 | 100±1 | 89±5 | 101±2 | 97±1 | 51±6 |
| CaMKKβ | 99±5 | 86±1 | 88±2 | 13±6 | 94±1 | 96±3 | 89±1 | 90±2 | 72±6 | 103±7 | 76±2 | 96±2 | 88±3 | 63±3 |
| CaMK1 | 86±6 | 91±1 | 84±6 | 99±3 | 86±9 | 90±3 | 91±0 | 83±3 | 63±3 | 83±6 | 47±1 | 77±9 | 53±3 | 89±2 |
| SmMLCK | 85±1 | 93±1 | 95±1 | 94±12 | 85±14 | 92±9 | 91±3 | 78±0 | 61±5 | 81±7 | 72±1 | 85±8 | 84±1 | 68±5 |
| PHK | 104±3 | 75±5 | 68±7 | 55±6 | 80±5 | 85±7 | 91±0 | 85±1 | 75±1 | 80±6 | 78±4 | 120±4 | 90±2 | 79±2 |
| CHK1 | 66±6 | 85±5 | 73±3 | 62±7 | 86±8 | 90±6 | 93±2 | 97±8 | 96±2 | 96±8 | 106±0 | 89±8 | 89±5 | 85±8 |
| CHK2 | 92±4 | 99±4 | 117±0 | 29±3 | 11±1 | 101±6 | 103±1 | 75±3 | 32±2 | 83±2 | 44±3 | 80±4 | 99±1 | 75±4 |
| GSK3β | 34±3 | 27±2 | 54±8 | 88±6 | 89±1 | 94±6 | 107±1 | 95±1 | 77±0 | 90±3 | 93±3 | 107±4 | 64±1 | 54±3 |
| CDK2-Cyclin A | 104±3 | 82±7 | 83±6 | 69±5 | 97±13 | 89±6 | 87±1 | 95±3 | 56±1 | 86±5 | 43±5 | 101±4 | 80±7 | 97±3 |
| PLK1 | 97±1 | 86±8 | 86±9 | 89±2 | 90±3 | 87±9 | 86±5 | 95±3 | 95±6 | 95±5 | 94±0 | 95±3 | 86±2 | 81±7 |
| Aurora B | 85±2 | 90±9 | 82±2 | 10±6 | 35±2 | 73±1 | 96±6 | 91±8 | 75±8 | 106±3 | 83±0 | 95±3 | 15±1 | 60±2 |
| Aurora C | 92±0 | 82±2 | 78±7 | 7±0 | 71±1 | 48±5 | 83±1 | 88±6 | 56±1 | 76±1 | 74±2 | 85±6 | 34±1 | 92±4 |
| AMPK | 80±0 | 91±3 | 75±3 | 10±6 | 87±11 | 102±9 | 113±1 | 82±6 | 73±2 | 86±3 | 97±2 | 82±2 | 89±4 | 97±4 |
| MARK3 | 98±2 | 66±2 | 63±4 | 44±7 | 92±10 | 128±5 | 126±8 | 67±2 | 30±1 | 72±6 | 55±2 | 122±9 | 83±0 | 92±1 |
| BRSK2 | 86±11 | 71±3 | 47±6 | 10±0 | 92±11 | 90±8 | 86±4 | 108±4 | 75±1 | 101±3 | 97±5 | 78±9 | 80±2 | 93±2 |
| MELK | 83±9 | 63±3 | 61±3 | 73±5 | 76±7 | 58±1 | 57±1 | 84±2 | 28±0 | 66±2 | 24±0 | 82±9 | 56±6 | 78±4 |
| CK1δ | 8±1 | 21±1 | 97±8 | 104±6 | 101±4 | 20±2 | 7±1 | 30±0 | 4±1 | 56±5 | 14±3 | 90±4 | 99±4 | 93±5 |
| CK2 | 98±2 | 103±3 | 96±5 | 109±1 | 92±2 | 99±4 | 108±4 | 88±0 | 87±0 | 80±7 | 92±3 | 93±3 | 95±3 | 68±3 |
| DYRK1A | 84±0 | 99±5 | 97±5 | 70±1 | 102±6 | 97±6 | 96±1 | 91±4 | 77±8 | 92±9 | 66±3 | 75±6 | 82±8 | 62±4 |
| DYRK2 | 97±2 | 102±8 | 82±4 | 85±8 | 100±1 | 85±1 | 101±1 | 95±2 | 97±7 | 94±2 | 112±6 | 92±1 | 81±4 | 84±9 |
| DYRK3 | 112±0 | 96±5 | 102±4 | 87±4 | 81±2 | 92±2 | 95±8 | 85±1 | 65±1 | 80±5 | 58±5 | 90±2 | 47±9 | 42±0 |
| NEK2a | 81±5 | 98±3 | 87±6 | 71±2 | 83±1 | 85±1 | 87±1 | 99±4 | 95±4 | 102±7 | 92±6 | 86±6 | 92±5 | 83±3 |
| NEK6 | 98±1 | 92±7 | 86±2 | 94±5 | 97±14 | 80±3 | 84±6 | 98±9 | 92±8 | 95±4 | 111±1 | 90±4 | 92±9 | 82±7 |
| NEK7 | 96±6 | 107±2 | 104±0 | 107±1 | 81±7 | 116±2 | 102±3 | 94±6 | 106±0 | 111±2 | 109±8 | 97±1 | 96±4 | 94±4 |
| IKKβ | 95±3 | 95±0 | 75±4 | 80±1 | 97±7 | 79±14 | 89±4 | 92±2 | 95±3 | 99±2 | 95±1 | 90±7 | 94±0 | 93±6 |
| PIM1 | 82±4 | 85±4 | 80±4 | 95±9 | 82±0 | 89±8 | 116±3 | 105±6 | 91±5 | 104±4 | 95±1 | 96±7 | 72±3 | 13±0 |
| PIM2 | 97±3 | 107±2 | 124±6 | 95±8 | 85±6 | 111±15 | 100±1 | 100±5 | 105±1 | 101±4 | 104±2 | 93±8 | 98±2 | 5±0 |
| PIM3 | 81±4 | 92±4 | 60±4 | 88±2 | 81±8 | 91±10 | 90±2 | 99±4 | 83±5 | 99±2 | 87±3 | 81±3 | 65±8 | 8±1 |
| SRPK1 | 91±2 | 85±0 | 95±1 | 29±1 | 69±1 | 93±4 | 86±9 | 94±8 | 89±8 | 100±4 | 93±1 | 87±3 | 88±5 | 79±4 |
| MST2 | 86±5 | 86±3 | 88±7 | 7±8 | 87±4 | 70±3 | 80±4 | 46±4 | 10±4 | 45±3 | 10±1 | 84±2 | 91±4 | 40±4 |
| EF2K | 98±0 | 92±5 | 92±3 | 108±2 | 89±1 | 127±7 | 106±7 | 98±1 | 100±2 | 99±2 | 105±2 | 106±0 | 99±1 | 105±3 |
| HIPK2 | 86±6 | 97±5 | 100±2 | 71±3 | 88±1 | 86±8 | 100±4 | 100±2 | 85±2 | 96±4 | 72±3 | 106±8 | 15±1 | 38±5 |
| HIPK3 | 96±1 | 109±1 | 104±1 | 95±1 | 83±13 | 102±3 | 112±2 | 101±3 | 105±1 | 96±3 | 103±3 | 107±8 | 21±3 | 79±8 |
| PAK4 | 86±13 | 83±6 | 71±6 | 57±2 | 95±1 | 92±7 | 96±6 | 82±3 | 35±0 | 71±6 | 50±8 | 98±4 | 89±5 | 95±9 |
| PAK5 | 89±1 | 89±9 | 92±2 | 75±2 | 81±2 | 87±3 | 94±7 | 73±9 | 25±4 | 74±0 | 47±1 | 96±2 | 89±3 | 97±1 |
| PAK6 | 83±10 | 84±2 | 83±0 | 80±0 | 102±12 | 96±2 | 98±5 | 94±6 | 57±6 | 89±1 | 66±6 | 107±1 | 94±2 | 98±8 |
| Src | 73±3 | 79±6 | 93±4 | 4±1 | 12±1 | 4±1 | 9±1 | 20±6 | 4±2 | 30±7 | 7±0 | 84±5 | 44±1 | 74±8 |
| Lck | 56±4 | 44±2 | 51±8 | 11±2 | 11±1 | 6±1 | 10±0 | 30±0 | 6±1 | 18±3 | 4±0 | 10±2 | 27±0 | 63±1 |
| CSK | 69±0 | 70±2 | 70±1 | 85±1 | 12±3 | 39±1 | 26±0 | 32±1 | 6±1 | 36±2 | 6±2 | 68±3 | 63±3 | 92±11 |
| RIP2 | 7±1 | 7±1 | 101±2 | ND | 9±1 | 9±1 | 7±1 | 10±1 | 6±1 | 14±1 | 5±1 | 92±5 | 26±2 | 35±2 |
| GAK | 25±2 | 0±1 | 94±1 | ND | 79±3 | 12±1 | 20±1 | 57±8 | 6±3 | 43±6 | 6±5 | 83±7 | 90±8 | 32±3 |
| c-Raf | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 0±1 | 1±1 | 24±1 |
| B-Raf | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 10±1 | 30±1 | 16±1 |
BIRB 0796 is a more potent inhibitor of p38α and p38β MAPKs than is SB 203580. It interacts with p38α MAPK in a manner distinct from that exhibited by SB 203580/SB 202190, and its binding induces a slow conformational change that locks the protein into an inactive conformation. Thus the potency of BIRB 0796 increases with the period of preincubation with the inhibitor [29]. In contrast with SB 203580 or SB 202190, we find that BIRB 0796 does not inhibit CK1δ, GSK3β, RIP2 or GAK in vitro (Table 1). However, unlike SB 203580/SB202190, BIRB 0796 also inhibits p38γ MAPK, p38δ MAPK and JNK2α2 (Table 1). As judged by suppression of the phosphorylation of well-established substrates, BIRB 0796 inhibits p38α MAPK activity completely when added to the culture medium at only 0.1 μM, but at 1 μM it also inhibits p38γ MAPK. Thus substrates for p38γ MAPK (and perhaps p38δ MAPK) can be identified as proteins whose phosphorylation is unaffected at 0.1 μM BIRB 0796, but inhibited at 1 μM BIRB 0796 [30]. Although BIRB 0796 is a potent inhibitor of JNK2 in vitro, it does not affect the phosphorylation of JNK substrates (c-Jun and ATF2) in cells at the low (0.1 μM) concentration that abolishes p38α MAPK activity in cells, because JNK1 is the dominant isoform that phosphorylates c-Jun and activates the AP1 transcription factor in the cells that have been studied so far [31,32].
We have used BIRB 0796 extensively to study the role of p38α MAPK and p38β MAPK in cell-based assays and we recommend that it be used in parallel with SB 203580 or SB 202190 when assessing the physiological roles of these protein kinases.
Src family kinase inhibitors (SU 6656, PP1, PP2 and Src inhibitor 1)
The compound SU 6656 is reported to be a potent inhibitor of Src family members [33]. In the present study we found that it inhibited AMPK, BRSK2 and MST2 with similar potency to its inhibition of Src and Lck, and it inhibited Aurora B and C, even more potently than Src and Lck in vitro (Tables 1 and 2). SU 6656 also inhibited other protein kinases, such as CaMKKα, CaMKKβ, CHK2 and SRPK1 (Table 1). These findings indicate that results obtained by using SU 6656 should be interpreted with caution.
Table 2. Potencies of compounds developed as Raf and Src inhibitors towards a variety of protein kinases.
IC50 values were determined from assays carried out at ten different inhibitor concentrations.
| Compound | Protein kinase | IC50 (μM) | [ATP] in assay (μM) |
|---|---|---|---|
| SU 6656 | Src | 0.10 | 50 |
| Lck | 0.15 | 50 | |
| Aurora B | 0.019 | 20 | |
| Aurora C | 0.017 | 5 | |
| BRSK2 | 0.10 | 50 | |
| MST2 | 0.11 | 50 | |
| AMPK | 0.11 | 50 | |
| Src-I1 | Src | 0.18 | 50 |
| RIP2 | 0.026 | 100 | |
| PP1 | Src | 0.053 | 50 |
| Lck | 0.040 | 50 | |
| RIP2 | 0.026 | 100 | |
| CK1δ | 0.17 | 20 | |
| CSK | 0.64 | 20 | |
| PP2 | Src | 0.036 | 50 |
| Lck | 0.031 | 50 | |
| RIP2 | 0.019 | 100 | |
| CK1δ | 0.041 | 20 | |
| NA-PP1 | JNK1[M108A] | 0.27 | 20 |
| JNK1[M108G] | 0.62 | 20 | |
| Src | 0.34 | 50 | |
| Lck | 0.66 | 50 | |
| CSK | 1.97 | 20 | |
| RIP2 | 0.12 | 100 | |
| PKD1 | 0.90 | 50 | |
| CKI | 0.15 | 20 | |
| NM-PP1 | JNK1[M108A] | 0.25 | 20 |
| JNK1[M108G] | 0.14 | 20 | |
| Src | 0.62 | 50 | |
| Lck | 0.46 | 50 | |
| CSK | 0.51 | 20 | |
| RIP2 | 0.13 | 100 | |
| PKA | 0.50 | 20 | |
| PKD1 | 0.25 | 50 | |
| ZM 336372 | c-RAF | 0.031 | 100 |
| B-RAF | 0.23 | 100 | |
| Lck | 0.57 | 50 | |
| RIP2 | >100 | 100 | |
| BAY 439006 | c-RAF | 0.37 | 100 |
| B-RAF | 7.3 | 100 | |
| p38αMAPK | 3.2 | 100 | |
| RIP2 | 3.6 | 100 | |
| GW 5074 | c-RAF | 0.17 | 100 |
| B-RAF | 0.19 | 100 | |
| PIM1 | 0.17 | 20 | |
| PIM2 | 0.07 | 5 | |
| PIM3 | 0.10 | 20 |
The related pyrazolopyrimidines PP1 and PP2 have been used widely to suggest physiological roles for Src family protein kinases, although they do not discriminate between different members of this family [34,35]. They also inhibit other protein tyrosine kinases, such as Eph-A2 and FGF-R1 (results not shown). In our assays, PP1 and PP2 inhibited Src and the closely related Lck with IC50 values of 50 nM, whereas CSK, p38α MAPK and CK1δ were inhibited with 3–10-fold lower potency. Interestingly, we found that RIP2 was inhibited even more potently than were Src or Lck (Tables 1 and 2), and we have recently exploited this finding to identify novel roles for RIP2 in cells [36].
Another compound, termed Src-I1, was found to be a potent inhibitor of Src (Table 2), but also inhibited other Src family members, such as Lck, Csk (Table 1) and Yes (results not shown) with similar potency to Src, and RIP2 with even greater potency (Table 2). In addition, it inhibited CHK2 with similar potency to Src, and Aurora B with slightly lower potency (Table 1). However, in contrast with PP1 and PP2, it did not inhibit p38α/p38β MAPKs or CK1δ (Table 1). We therefore recommend that PP1 or PP2 be used in parallel with Src-I1 to assess the physiological roles of the Src family of protein tyrosine kinases.
PP1 derivatives NM-PP1 and NA-PP1
A significant subset of protein kinases, including Src, Lck, p38α/p38β MAPKs, GAK, RIP2 and a number of receptor tyrosine kinases, possess a threonine residue at the so-called ‘gatekeeper’ site. This creates a hydrophobic pocket near the ATP-binding site, which underlies the sensitivity of these enzymes to compounds such as PP1/PP2 and/or SB 203580. By contrast, these compounds do not inhibit most protein kinases because they possess a bulky hydrophobic residue at this position. For example, in v-Src, the virally encoded form of Src, the threonine residue is replaced by isoleucine, explaining why this oncogene product is insensitive to PP1/PP2 [35]. However, by mutating the residue at the gatekeeper site to threonine or other amino acids with even smaller side chains (serine, alanine or glycine), it is possible to convert protein kinases into forms that can be potently inhibited by PP1, PP2 or SB 203580. Conversely, the mutation of the gatekeeper threonine residue into an amino acid with a larger side chain converts these protein kinases into SB203580-insensitive forms [37–39].
Recently, ‘knock-in’ mice have been generated that express a mutated form of JNK in which the gatekeeper methionine residue has been changed to glycine [31,32]. In contrast with wild-type JNK, the mutated JNK can be inhibited by modified PP1 derivatives, such as NA-PP1 and NM-PP1. Potentially, this is a powerful way of studying the physiological roles of protein kinases, because the mutated kinase possesses an activity similar to that of the wild-type enzyme, but can be inhibited rapidly and reversibly by adding NA-PP1 or NM-PP1 to the culture medium. However, the general applicability of this approach depends, in part, on the selectivity with which NA-PP1 and NM-PP1 inhibit the mutant protein kinases compared with the other wild-type protein kinases that are expressed endogenously in the same cells and tissues. We therefore examined the specificities of NA-PP1 and NM-PP1 against our extended panel of kinases.
The specificities of NA-PP1 and NM-PP1 were similar to those exhibited by PP1 and PP2, these compounds inhibiting RIP2, GAK, CK1 and p38α/β MAPK, as well as Src, Lck and Csk (Table 1) and other protein-tyrosine kinases such as Eph-A2 and FGF-R1 (results not shown). Additionally, we found that NA-PP1 and NM-PP1 inhibited PKD1 and MST2, whereas NM-PP1 also inhibits PKA (Tables 1 and 2). We also found that the concentrations of NA-PP1 and NM-PP1 required to inhibit the gatekeeper mutants of JNK1 (JNK1[M108A] and JNK1[M108G]) were similar to those required to inhibit the Src family kinases RIP2 and PKD (Table 2). Wild-type JNK1 was not inhibited by NA-PP1 or NM-PP1 (Table 1).
These findings suggest that caution may be needed in interpreting experiments performed using cells and tissues from mice that express the gatekeeper mutants of protein kinases (sensitized to inhibition by NA-PP1/NM-PP1) instead of the wild-type enzymes. Although control experiments can be carried out using cells/tissues from wild-type mice or knock-out mice that do not express the protein kinase, to check for ‘off-target’ effects of NA-PP1 and NM-PP1, it is often necessary to inhibit protein kinases in two different signalling pathways in order to suppress the phosphorylation of a particular protein or biological process. For example, the combined inhibition of MKK1 and p38α MAPK is needed to suppress the phosphorylation of CREB (cAMP-response-element-binding protein) induced by EGF or UV-C radiation [40,41], whereas the combined inhibition of PI3K [phosphatidylinositol (phosphoinositide) 3-kinase] and MKK1 is needed to prevent the EGF-stimulated phosphorylation of GSK3 [41]. It is therefore possible that the effects of NA-PP1/NM-PP1 on cells do not always result from the inhibition of the gatekeeper mutant kinase alone, but may result from the combined inhibition of the mutant kinase and one or more other intracellular protein kinases, such as Src family members RIP2 and PKD1, which are inhibited by these compounds at similar concentrations.
Raf inhibitors ZM 336372, BAY 439006 and GW 5074
The Raf isoforms lie at the head of the classical growth-factor-stimulated MAP kinase cascade that plays a key role in stimulating cells to proliferate or differentiate. Activating mutations in B-Raf occur in many cancers and with high frequency in malignant melanoma. ZM 336372 was originally developed as a c-Raf inhibitor. Like p38α MAPK and p38β MAPK, Raf possesses a threonine residue at the gatekeeper site, explaining why ZM 336372 inhibits p38α/β MAPKs and why SB 203580 inhibits Raf. Thus the mutation of Thr106 in p38α MAPK to methionine makes it insensitive to both ZM 336372 and SB 203580 [25,42]. Here we extended the specificity of ZM 336372 to 70 protein kinases, which established that it does not inhibit other protein kinases tested significantly, apart from three that possess a threonine residue at the gatekeeper site (p38α MAPK, p38β MAPK and Lck) (Tables 1 and 2).
Despite being a potent and specific inhibitor of Raf, ZM 336372 does not prevent the growth-factor- or phorbol-ester-induced activation of MKK1 or ERK1/ERK2 and, unlike inhibitors of MKK1, it does not reverse the phenotype of Ras- or Raf-transformed cell lines [42]. This appears to be explained by a feedback control loop in which Raf efficiently prevents its own activation, such that the inhibition of Raf by ZM 336372 is always counterbalanced by an equivalent activation, resulting from the suppression of this feedback loop [42]. These findings have highlighted a problem in targeting Raf for the development of anti-cancer drugs.
BAY 439006 was also developed initially as a Raf inhibitor [43] and, in the present study, we found that its specificity resembles that of ZM 336372. Thus, like ZM 336372, BAY 439006 also inhibits p38α MAPK, p38β MAPK, Src and Lck. However, unlike ZM 336372, BAY 439006 also inhibits RIP2, Aurora kinases, HIPK2, HIPK3 and ERK8 (Tables 1 and 2). BAY 439006 (also called Nexavar) has been approved for the treatment of kidney cancer and gastrointestinal tumours that are resistant to Gleevec. Although originally believed to exert its anticancer effects by inhibiting Raf, more recent studies have demonstrated that it also inhibits a number of receptor tyrosine kinases that possess threonine at the gatekeeper site, and that inhibition of these targets, rather than Raf, is likely to underlie its clinical efficacy [44]. Consistent with this, we have found that BAY 439006 inhibits FGF-R1 and Eph-A2 tyrosine kinases (results not shown).
GW 5074 is another inhibitor of Raf isoforms (Table 2). In the present study we found that this compound inhibited the three PIM isoforms more potently than Raf, and it also inhibited several other protein kinases, such as HIPK2, RIP2, GAK and MST2, with a potency comparable with that towards Raf (Tables 1 and 2).
In summary, the feedback-control mechanism by which Raf suppresses its own activation means that no compounds have yet been developed that convincingly prevent the activation of the classical MAPK cascade by inhibiting Raf, and inhibitors of MKK1 are being used for this purpose instead, as detailed below.
Inhibitors of MKK1 and its activation (U0126, PD 184352, PD 0325901 and PD 0325901-Cl)
MKK1 (also called MEK1) is activated by Raf in vivo and is also being targeted to develop anticancer drugs. PD 98059 [45] and U0126 [46] were the first compounds to be described that target MKK1 and the closely related MKK2 and have been exploited in thousands of subsequent studies. Although initially identified by their ability to inhibit a mutated form of MKK1 that possesses some constitutive activity in vitro, PD 98059 and U0126 are non-competitive inhibitors that appear to interact with the inactive unphosphorylated kinase more strongly than the active phosphorylated species and therefore exert their effects in cell-based assays by preventing the phosphorylation of MKK1 and/or the conformational transition that generates the activated enzyme [1,45]. More recently, additional non-competitive inhibitors of MKK1 with greater potency (PD 184352 [47] and PD 0325901 [5]) have been developed and have entered clinical trials as anti-cancer agents.
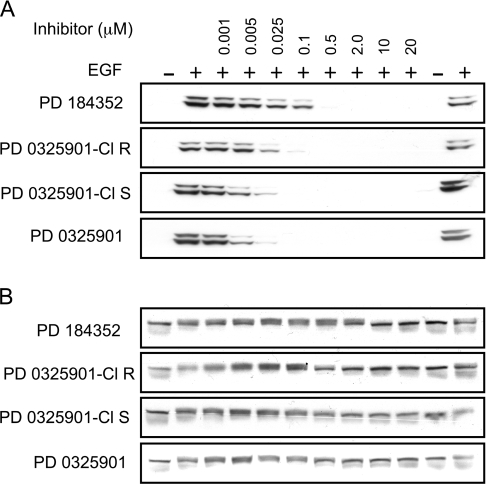
The specificities of U0126, PD 184352, PD 0325901 and the (S) stereoisomer of a closely related compound, termed here PD 0325901-Cl (Supplementary Figure S1) are compared in Table 3. PD 184352, PD 0325901 and PD 0325901-Cl inhibited the active phosphorylated form of MKK1 with IC50 values close to 1 μM in vitro, whereas U0126 inhibited activated MKK1 with about 10-fold lower potency. However, these non-competitive inhibitors suppressed the activation of ERK1/ERK2 (the substrates of MKK1) at much lower concentrations in cell-based assays, presumably because they bind even more strongly to the inactive unphosphorylated form of MKK1. We have reported previously that the EGF-induced activation of ERK1/ERK2 was completely suppressed at 10 μM U0126 or 1 μM PD 184352 in Swiss 3T3 cells [1]. In the present study, we found that PD 0325901 and the (S) and (R) isomers of PD 0325901-Cl were even more potent inhibitors than PD 184352. PD 0325901 and the (S) isomer of PD 0325901-Cl suppressed the activation of ERK1/ERK2 at 25 nM in EGF-stimulated HeLa cells, as compared with 0.5 μM for PD 184352 in parallel experiments. The (R) isomer of PD 0325901-Cl was a slightly less potent inhibitor than the (S) isomer (Figure 1A). At these concentrations, no other protein kinases in our panel were inhibited and, even at 10 μM, only a few protein kinases were inhibited slightly (Table 3).
Table 3. Specificities of inhibitors of MKK1, Raf and GSK3.
The concentrations of compounds used in the assays are indicated below each molecule and the results shown are presented as the percentage activity remaining in the presence of inhibitor as compared with control incubations with inhibitor omitted (averages of duplicate determinations). Each experiment was repeated two or three times with similar results. Further details of the assays are given in the Materials and methods section. Important values are highlighted in boldface type. ND, not determined.
| Percentage activity remaining | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kinase | U0126 (10 μM) | PD 184352 (10 μM) | PD 0325901 (10 μM) | PD 0325901-Cl (10 μM) | BI-D1870 (0.1 μM) | SL0101 (10 μM) | FMK (3 μM) | CT 99021 (1 μM) | AR-A0-14418 (10 μM) | SB 216763 (2 μM) | SB 415286 (10 μM) | Alsterpaullone (1 μM) | Kenpaullone (1 μM) | LiCl (10 mM) |
| MKK1 | 24±1 | 12±1 | 5±1 | 6±5 | 98±4 | 88±7 | 80±8 | 71±3 | 77±9 | 71±6 | 28±3 | 86±3 | 88±3 | 83±8 |
| ERK1 | 72±5 | 86±0 | 90±1 | 104±5 | 115±2 | 98±5 | 92±6 | 77±2 | 101±1 | 99±1 | 94±9 | 95±0 | 86±7 | 101±6 |
| ERK2 | 68±3 | 83±6 | 91±6 | 98±7 | 85±1 | 98±9 | 79±5 | 90±6 | 95±8 | 97±7 | 96±9 | 79±5 | 88±9 | 71±7 |
| JNK1 | 90±2 | 92±1 | 101±12 | 90±1 | 88±6 | 95±10 | 81±3 | 85±8 | 81±2 | 106±8 | 96±1 | 82±5 | 89±3 | 81±4 |
| JNK2 | 73±4 | 110±4 | 96±7 | 96±9 | 115±12 | 94±7 | 83±5 | 88±3 | 72±5 | 85±6 | 88±4 | 81±4 | 83±2 | 85±8 |
| JNK3 | 98±4 | 108±9 | 117±1 | 82±0 | 97±10 | 98±2 | 89±4 | 71±4 | 92±0 | 88±1 | 90±6 | 78±0 | 85±0 | 77±4 |
| p38α MAPK | 77±6 | 86±2 | 105±7 | 93±3 | 88±13 | 94±8 | 72±2 | 82±9 | 110±9 | 77±11 | 72±6 | 109±1 | 94±5 | 100±8 |
| P38β MAPK | 91±9 | 96±5 | 96±5 | 107±2 | 100±4 | 98±2 | 85±4 | 87±4 | 103±8 | 85±10 | 89±7 | 86±2 | 95±1 | 102±2 |
| p38γ MAPK | 90±1 | 87±2 | 103±11 | 99±1 | 111±4 | 103±8 | 92±7 | 101±7 | 89±1 | 84±9 | 98±7 | 87±5 | 86±5 | 90±1 |
| p38δ MAPK | 93±7 | 93±2 | 98±14 | 80±1 | 105±8 | 88±10 | 103±9 | 100±6 | 81±3 | 81±6 | 82±2 | 86±0 | 93±7 | 78±13 |
| ERK8 | 67±3 | 94±2 | 67±12 | 104±7 | 82±6 | 89±9 | 58±4 | 91±1 | 37±3 | 12±1 | 13±3 | 87±5 | 75±6 | 75±5 |
| RSK1 | 83±6 | 124±11 | 85±11 | 99±2 | 1±1 | 23±1 | 92±6 | 65±8 | 79±4 | 50±3 | 31±2 | 67±3 | 83±3 | 80±8 |
| RSK2 | 81±1 | 92±4 | 81±3 | 94±3 | 2±0 | 30±5 | 74±2 | 88±6 | 84±1 | 67±7 | 29±1 | 73±7 | 83±8 | 57±8 |
| PDK1 | 77±3 | 79±4 | 96±1 | 84±1 | 94±8 | 103±8 | 89±10 | 86±1 | 74±1 | 92±6 | 62±2 | 71±5 | 62±3 | 84±7 |
| PKBα | 76±8 | 87±7 | 94±1 | 92±1 | 98±1 | 89±8 | 82±3 | 106±1 | 95±2 | 88±9 | 92±1 | 88±1 | 95±6 | 77±5 |
| PKBβ | 98±1 | 80±0 | 116±6 | 91±5 | 102±15 | 99±4 | 97±4 | 80±6 | 86±2 | 96±9 | 85±10 | 88±1 | 90±7 | 77±6 |
| SGK1 | 72±6 | 101±6 | 75±8 | 91±3 | 112±7 | 110±11 | 107±8 | 79±4 | 88±4 | 100±5 | 82±6 | 85±1 | 80±0 | 119±7 |
| S6K1 | 93±5 | 89±1 | 106±8 | 92±1 | 119±8 | 103±8 | 33±2 | 84±2 | 75±2 | 86±11 | 81±2 | 93±1 | 114±2 | 127±14 |
| PKA | 93±5 | 87±0 | 108±7 | 70±3 | 89±8 | 92±7 | 85±7 | 89±0 | 68±4 | 88±6 | 80±8 | 79±1 | 63±4 | 76±5 |
| ROCK 2 | 81±6 | 86±0 | 82±4 | 100±7 | 84±14 | 109±4 | 86±7 | 79±4 | 83±2 | 101±7 | 79±4 | 87±5 | 42±6 | 109±14 |
| PRK2 | 85±9 | 83±5 | 101±4 | 81±7 | 97±5 | 107±7 | 83±3 | 93±1 | 85±2 | 81±7 | 43±7 | 61±6 | 58±5 | 86±8 |
| PKCα | 95±5 | 88±4 | 103±5 | 89±4 | 90±9 | 90±7 | 85±7 | 78±8 | 80±3 | 73±7 | 39±3 | 80±7 | 87±6 | 90±9 |
| PKCζ | 78±2 | 72±3 | 97±4 | 82±5 | 80±1 | 82±3 | 91±5 | 90±2 | 70±1 | 81±3 | 69±4 | 50±4 | 72±6 | 72±2 |
| PKD1 | 77±6 | 85±3 | 90±7 | 83±1 | 63±11 | 89±3 | 83±4 | 75±9 | 93±5 | 100±10 | 64±8 | 91±9 | 87±1 | 95±3 |
| MSK1 | 85±7 | 91±5 | 87±10 | 90±1 | 125±5 | 101±1 | 75±3 | 83±9 | 81±4 | 69±2 | 62±5 | 82±6 | 88±1 | 85±6 |
| MNK1 | 78±2 | 92±9 | 94±2 | 92±4 | 87±9 | 82±0 | 42±2 | 81±6 | 80±3 | 85±6 | 77±8 | 74±1 | 85±3 | 53±8 |
| MNK2 | 85±8 | 99±3 | 104±3 | 106±3 | 96±12 | 97±6 | 65±6 | 85±9 | 95±6 | 94±3 | 91±8 | 90±4 | 102±1 | 56±3 |
| MAPKAP-K2 | 95±2 | 81±5 | 112±8 | 106±8 | 89±15 | 95±4 | 94±10 | 110±4 | 103±2 | 82±2 | 85±5 | 95±4 | 92±6 | 76±7 |
| MAPKAP-K3 | 97±15 | 84±8 | 114±12 | 97±1 | 108±13 | 112±12 | 92±6 | 71±8 | 82±4 | 103±2 | 99±6 | 70±5 | 97±2 | 92±12 |
| PRAK | 92±2 | 82±5 | 97±7 | 81±1 | 89±15 | 82±3 | 97±5 | 76±7 | 67±5 | 90±10 | 81±5 | 76±0 | 80±1 | 75±4 |
| CaMKKα | 99±11 | 77±1 | 80±1 | 90±1 | 92±8 | 116±9 | 70±12 | 89±3 | 79±5 | 85±4 | 53±6 | 80±3 | 74±5 | ND |
| CaMKKβ | 92±6 | 90±1 | 97±6 | 103±1 | 73±13 | 108±7 | 49±3 | 84±2 | 73±9 | 82±4 | 29±3 | 68±3 | 61±2 | 104±8 |
| CaMK1 | 69±4 | 69±4 | 109±8 | 103±1 | 103±9 | 105±8 | 77±9 | 84±1 | 76±3 | 96±10 | 61±3 | 54±2 | 88±6 | 79±4 |
| SmMLCK | 95±5 | 105±3 | 111±2 | 82±1 | 107±4 | 109±4 | 82±4 | 83±1 | 91±3 | 60±6 | 65±8 | 91±4 | 84±1 | 43±6 |
| PHK | 94±7 | 98±2 | 102±11 | 83±6 | 114±14 | 97±5 | 93±5 | 75±1 | 90±4 | 87±5 | 84±3 | 11±1 | 18±1 | 60±2 |
| CHK1 | 83±3 | 113±1 | 97±11 | 88±9 | 73±0 | 82±1 | 88±4 | 97±2 | 83±2 | 87±6 | 60±5 | 64±4 | 72±5 | 72±5 |
| CHK2 | 98±3 | 95±8 | 102±5 | 87±8 | 96±3 | 99±2 | 81±9 | 93±4 | 93±3 | 93±4 | 69±10 | 13±4 | 28±2 | 59±8 |
| GSK3β | 82±5 | 78±6 | 100±15 | 76±0 | 77±5 | 87±11 | 65±6 | 1±2 | 3±0 | 2±2 | 4±1 | 3±0 | 3±1 | 30±4 |
| CDK2-Cyclin A | 86±1 | 88±8 | 90±5 | 94±7 | 84±0 | 106±5 | 80±7 | 53±3 | 23±3 | 52±5 | 18±1 | 10±2 | 14±0 | 100±1 |
| PLK1 | 92±7 | 82±0 | 105±7 | 84±8 | 5±2 | 112±11 | 82±5 | 41±2 | 78±0 | 62±6 | 85±7 | 81±5 | 90±1 | 75±4 |
| Aurora B | 86±3 | 70±1 | 87±3 | 80±3 | 22±5 | 45±7 | 59±3 | 85±9 | 71±4 | 87±3 | 64±3 | 78±3 | 49±4 | 57±2 |
| Aurora C | 88±11 | 96±9 | 118±6 | 80±3 | 76±4 | 77±2 | 75±6 | 83±7 | 84±3 | 79±7 | 70±4 | 76±4 | 83±1 | 60±2 |
| AMPK | 76±12 | 108±3 | 89±6 | 114±4 | 108±6 | 88±0 | 79±4 | 85±4 | 72±1 | 67±5 | 33±4 | 73±6 | 94±1 | 103±7 |
| MARK3 | 95±5 | 98±2 | 105±5 | 101±7 | 79±5 | 113±7 | 80±3 | 91±9 | 75±5 | 76±3 | 29±2 | 63±5 | 36±5 | 89±6 |
| BRSK2 | 87±12 | 82±5 | 97±7 | 87±4 | 78±9 | 80±5 | 53±6 | 67±1 | 76±4 | 83±2 | 38±3 | 37±4 | 41±0 | 81±7 |
| MELK | 83±11 | 82±3 | 107±1 | 73±3 | 40±5 | 102±6 | 65±8 | 45±5 | 75±4 | 78±2 | 72±5 | 54±2 | 55±6 | 82±5 |
| CK1δ | 90±10 | 100±1 | 103±0 | 106±4 | 81±5 | 88±1 | 63±6 | 85±4 | 89±6 | 78±5 | 68±4 | 98±4 | 98±1 | 55±10 |
| CK2 | 99±4 | 90±8 | 100±7 | 100±7 | 97±4 | 101±7 | 92±10 | 72±2 | 79±1 | 89±3 | 97±1 | 88±1 | 78±5 | 86±4 |
| DYRK1A | 69±4 | 80±2 | 87±11 | 84±1 | 96±9 | 80±7 | 86±11 | 81±9 | 50±6 | 8±4 | 30±3 | 83±1 | 80±4 | 89±4 |
| DYRK2 | 84±3 | 76±2 | 99±1 | 110±6 | 91±1 | 89±4 | 91±8 | 87±2 | 81±7 | 92±7 | 83±7 | 87±3 | 73±5 | 94±6 |
| DYRK3 | 77±4 | 104±2 | 88±8 | 79±1 | 91±3 | 85±3 | 68±7 | 66±2 | 71±1 | 79±5 | 84±5 | 93±9 | 81±4 | 88±8 |
| NEK2a | 89±7 | 98±1 | 95±10 | 99±2 | 98±10 | 101±8 | 95±4 | 83±1 | 81±3 | 74±7 | 86±6 | 92±2 | 87±1 | 74±8 |
| NEK6 | 90±3 | 90±6 | 106±9 | 92±4 | 115±8 | 109±13 | 90±4 | 89±5 | 91±1 | 74±4 | 74±1 | 86±1 | 93±1 | 53±12 |
| NEK7 | 97±6 | 93±1 | 120±6 | 99±2 | 108±0 | 105±1 | 92±8 | 85±0 | 96±3 | 85±5 | 86±9 | 92±5 | 97±7 | 73±6 |
| IKKβ | 118±7 | 92±0 | 95±5 | 89±1 | 106±11 | 100±2 | 92±8 | 88±9 | 75±3 | 99±2 | 84±5 | 90±2 | 76±4 | 88±10 |
| PIM1 | 87±0 | 75±3 | 86±3 | 75±1 | 76±2 | 71±3 | 61±6 | 70±2 | 83±4 | 56±10 | 37±2 | 81±5 | 49±0 | 68±1 |
| PIM2 | 101±3 | 92±1 | 104±5 | 90±1 | 100±8 | 95±1 | 85±10 | 63±1 | 92±5 | 81±4 | 46±3 | 91±5 | 80±1 | 120±6 |
| PIM3 | 63±8 | 66±4 | 62±5 | 46±2 | 65±3 | 49±2 | 56±6 | 64±4 | 69±1 | 29±7 | 46±8 | 78±8 | 28±0 | 97±2 |
| SRPK1 | 102±6 | 94±4 | 95±6 | 100±5 | 94±7 | 96±5 | 85±1 | 81±6 | 74±6 | 16±1 | 54±6 | 79±7 | 92±4 | 85±12 |
| MST2 | 96±5 | 89±1 | 99±1 | 86±5 | 41±4 | 99±2 | 76±4 | 79±7 | 80±7 | 50±7 | 14±2 | 53±3 | 41±2 | 94±5 |
| EF2K | 116±5 | 97±0 | 103±4 | 99±4 | 88±0 | 86±5 | 97±8 | 99±1 | 98±5 | 98±5 | 94±2 | 87±4 | 110±1 | 58±6 |
| HIPK2 | 97±3 | 151±5 | 110±2 | 98±3 | 103±7 | 72±9 | 79±11 | 84±4 | 53±3 | 15±1 | 63±2 | 77±2 | 81±9 | 66±6 |
| HIPK3 | 87±7 | 95±6 | 122±2 | 102±1 | 104±5 | 110±7 | 79±9 | 89±1 | 88±1 | 41±2 | 85±6 | 95±1 | 96±0 | 50±8 |
| PAK4 | 94±4 | 82±3 | 99±11 | 90±1 | 81±11 | 95±5 | 91±1 | 74±1 | 66±2 | 84±13 | 52±4 | 33±0 | 8±0 | 81±6 |
| PAK5 | 86±2 | 98±9 | 92±7 | 75±3 | 90±5 | 86±6 | 88±6 | 80±3 | 76±1 | 85±4 | 67±2 | 40±5 | 13±2 | 81±9 |
| PAK6 | 85±11 | 100±1 | 129±4 | 77±7 | 102±9 | 127±9 | 93±5 | 84±1 | 74±2 | 90±5 | 74±1 | 64±9 | 39±5 | 89±2 |
| Src | 84±6 | 90±5 | 95±1 | 97±5 | 96±3 | 105±3 | 10±1 | 83±2 | 81±1 | 83±10 | 77±7 | 61±2 | 38±1 | 62±4 |
| Lck | 84±4 | 85±2 | 92±10 | 88±1 | 85±0 | 88±0 | 10±1 | 88±3 | 80±1 | 77±5 | 66±3 | 52±4 | 29±3 | 76±10 |
| CSK | 88±12 | 81±2 | 117±3 | 82±2 | 99±9 | 105±6 | 55±7 | 66±1 | 82±2 | 88±6 | 89±2 | 83±9 | 77±5 | 89±2 |
| IKKϵ | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 51±8 |
| TBK1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 54±5 |
Figure 1. Effect of MKK1/MKK5 inhibitors on the activation of ERK1/ERK2 and ERK5.
HeLa cells were serum-starved for 16 h and incubated for a further 1 h in the presence (+) or absence (−) of PD184352, PD 0325901-Cl [(R) steroisomer], PD 0325901 [(S) steroisomer] or PD 0325901 at the concentrations indicated. The cells were then stimulated for 10 min with 100 ng/ml EGF, lysed, and 30 μg of the extract protein was denatured in SDS, subjected to SDS/PAGE, transferred to nitrocellulose membranes and immunoblotted with an antibody recognizing phosphorylated ERK1 and ERK2 in (A) and immunoblotted with an antibody that recognizes all forms of ERK5 equally well in (B).
PD 98059 and U0126 have been reported to inhibit MKK5, a protein kinase closely related to MKK1, with similar potency to MKK1 [49]. Thus these compounds also prevent the activation of ERK5, the physiological substrate of MKK5. We have reported that concentrations of PD 184352 which block the activation of ERK1/ERK2 in cells (1–2 μM) do not affect the activation of ERK5, and that higher concentrations (10–20 μM) are needed to prevent the activation of ERK5 in cells [50]. Here we show that PD 0325901 and PD 0325901-Cl also prevent the activation of ERK1/ERK2 in cells at concentrations that do not affect the activation of ERK5, as judged by their failure to prevent the EGF-induced phosphorylation of ERK5, measured by a decrease in electrophoretic mobility. However, these compounds blocked the activation of ERK5 when included in the culture medium at concentrations of 2 μM or higher (Figure 1B).
In summary, PD 184352 and PD 0325901/PD 0325901-Cl are both extremely potent and selective inhibitors of MKK1 (and the closely related MKK2) in cell-based assays and can also be used to suppress the activation of ERK5. Physiological substrates for ERK5 can be identified as proteins whose phosphorylation in cells is unaffected by 0.1 μM PD 0325901, but prevented by 2 μM PD 0325901, or as proteins whose phosphorylation is unaffected by 1–2 μM PD 184352, but suppressed 10-20 μM PD 184352. We recommend that PD 184352 or PD 0325901 be used to inhibit MKK1 in cells. The structurally unrelated U0126 can be used to check the results.
RSK inhibitors BI-D1870, SL0101 and FMK
The RSK isoforms are activated by ERK1/ERK2 and are the most downstream kinases of the classical MAPK cascade. We have recently described BI-D1870 as a relatively specific nanomolar inhibitor of RSK isoforms and exploited it to identify physiological substrates and roles for RSK in cells [51]. BI-D1870 was originally developed in a programme to identify inhibitors of PLKs, and it also inhibits PLK1 with slightly lower potency than RSK isoforms, whereas Aurora B, MELK, PIM3 and MST2, were inhibited with 10–100-fold lower potency and other protein kinases tested were unaffected [51] (Table 3).
In the present study we compared BI-D1870 with SL0101 [52] and FMK [53], two other recently described inhibitors of RSK (Table 3). These experiments revealed that SL0101 was also a relatively specific inhibitor of RSK isoforms, but much less potent than BI-D1870 (Table 1). SL0101 inhibited Aurora B, PIM1 and PIM3 with slightly lower potency than RSK1/RSK2, but other protein kinases in the panel were unaffected, including PLK1.
RSK isoforms are unusual in possessing two protein kinase domains in the same polypeptide. ERK1/ERK2 phosphorylate and activate the C-terminal kinase domain, which then activates the N-terminal kinase domain, enabling the N-terminal kinase domain to phosphorylate other proteins. FMK is an irreversible inhibitor that covalently modifies the C-terminal kinase domain of RSK. It therefore prevents the activation of the N-terminal kinase domain of RSK by the C-terminal kinase domain, but does not affect the activity of the N-terminal domain, explaining why the active forms of RSK1 and RSK2 are not inhibited by FMK in vitro (Table 3 and [53]). This contrasts with BI-D1870 and SL0101, which inhibit the N-terminal kinase domain. In the present study we found that FMK inhibited relatively few protein kinases in the panel, although it did inhibit protein tyrosine kinases, such as Src, Lck, Yes and Eph-A2, as well as S6K1 (Tables 3 and 5).
Table 5. Relative potencies of small-molecule kinase inhibitors towards different protein kinases.
IC50 values were determined from assays carried out at ten different inhibitor concentrations.
| Compound | Protein kinase | IC50 (μM) | ATP in assay (μM) |
|---|---|---|---|
| FMK | Src | 0.56 | 50 |
| Lck | 0.75 | 50 | |
| S6K1 | 0.95 | 20 | |
| Yes | 0.51 | 20 | |
| Eph-A2 | 0.76 | 50 | |
| Wortmannin | smMLCK | 5.0 | 50 |
| PLK1 | 1.3 | 5 | |
| LY 294002 | PLKI | 2.0 | 5 |
| PIM1 | 0.82 | 20 | |
| PIM3 | 1.4 | 20 | |
| D4476 | CK1 | 0.3 | 100 |
| PKD1 | 9.1 | 50 | |
| p38αMAPK | 5.8 | 50 | |
| Harmine | DYRK1A | 0.08 | 50 |
| DYRK2 | 0.9 | 50 | |
| DYRK3 | 0.8 | 50 | |
| PIM3 | 4.3 | 20 | |
| CK1 | 1.5 | 20 | |
| Roscovitine | CDK2 | 0.14 | 20 |
| PAK4 | 6.9 | 5 | |
| Purvalanol | CDK2 | 0.03 | 20 |
| PAK4 | 0.13 | 5 | |
| VX 680 | Aurora B | 0.04 | 100 |
| Aurora C | 0.07 | 100 | |
| Src | 0.34 | 50 | |
| MELK | 0.42 | 50 | |
| SU 6668 | Aurora B | 0.035 | 20 |
| Aurora C | 0.21 | 5 | |
| STO 609 | CaMKKβ | 0.01 | 20 |
| CaMKKα | 0.12 | 20 | |
| MNKI | 0.12 | 50 | |
| CK2 | 0.19 | 5 | |
| AMPK | 0.16 | 50 | |
| PIM2 | 0.11 | 5 | |
| PIM3 | 0.083 | 100 | |
| DYRK2 | 0.95 | 50 | |
| PS 1145 | IKKβ | 0.25 | 5 |
| PIMI | 1.1 | 20 | |
| PIM3 | 0.88 | 20 | |
| BMS 345541 | IKKβ | 2.6 | 5 |
| SC 514 | IKKβ | 2.0 | 5 |
| AS 601245 | JNK1 | 2.6 | 20 |
| JNK2 | 5.0 | 20 | |
| GSK3 | 0.04 | 20 | |
| PIM1 | 0.08 | 20 | |
| PIM3 | 0.03 | 5 | |
| DYRK2 | 0.3 | 50 | |
| CGP 57380 | MNKI | 0.87 | 50 |
| MNK2 | 1.6 | 50 | |
| CKI | 0.51 | 20 | |
| Aurora B | 2.5 | 20 | |
| DYRK3 | 3.2 | 5 | |
| SGK1 | 2.7 | 20 | |
| BRSK2 | 1.1 | 50 | |
| Lck | 2.5 | 50 |
In summary, we [51] and others [54] have found D1870 to be a useful inhibitor of RSK isoforms in cells and recommend it for this purpose, although it should be born in mind that PLKs will also be inhibited. SL0101 [52] (C. Watts, personal communication) and FMK [55] are also useful. FMK is the only known inhibitor of the C-terminal kinase domain of RSK and may therefore have a further use in preventing the phosphorylation of any proteins, besides the N-terminal kinase domain of RSK, that might be targeted by the C-terminal domain in cells. However, FMK would not inhibit RSK if the N-terminal kinase domain were activated by a mechanism that was independent of the C-terminal domain, as has recently been observed [54,55].
Inhibitors of GSK3 (CT 99021, AR-A0-144-18, SB 216763, SB 415286, alsterpaullone, kenpaullone and LiCl)
Inhibitors of GSK3 are being developed as potential drugs to treat diabetes, stroke, Alzheimer's and other diseases [56]. The compounds alsterpaullone, kenpaullone [2], CT 99021, AR-A0144-18, SB 216763 and SB 415286 [57] inhibit GSK3 at nanomolar concentrations. In the present study we found that CT 99021 was the most potent and specific inhibitor in vitro. It inhibited CDK2–cyclin A about 50-fold less potently and did not affect other protein kinases in the panel significantly at 1 μM (Tables 2 and 3). Apart from AR-A014418, the other four GSK3 inhibitors inhibited several other protein kinases in addition to CDK2–cyclin A. For example, SB 216763 inhibited ERK8, DYRK1A, PIM3, SRPK1 and HIPK2, SB 415286 inhibited MKK1, ERK8 and MST2 and several other protein kinases to a slightly lesser extent, kenpaullone inhibited PHK, CHK2, PAK4, PAK5, PIM3, Src and Lck, whereas alsterpaullone inhibited PHK and CHK2 (Table 3). SB 216763, SB 415286, kenpaullone and alsterpaullone also inhibited other protein kinases less strongly.
Lithium ions inhibit GSK3 in the millimolar range, and its effects in cell-based assays have been used to suggest physiological roles for this enzyme. In the present study we found that LiCl inhibited GSK3β activity in vitro more strongly than any of the other protein kinases tested. However, LiCl inhibited a number of other protein kinases with slightly lower potency than GSK3, including, MNK1, MNK2, smMLCK, PHK, CHK2, HIPK3, IKKϵ and TBK1 (Table 3).
In summary, we recommend using CT 99021 to inhibit GSK3 in cells, as it is the most potent and specific inhibitor available. When added to the cell culture medium at 1–2 μM, it completely prevents the phosphorylation of authentic GSK3 substrates such as NDRG1 (N-myc downstream-regulated gene 1) [57] and c-Jun at Thr239 (S. Morton and P. Cohen, unpublished work). Results obtained with CT 99021 can be checked by using one or more of the other GSK3 inhibitors.
Inhibitors of the PI3K superfamily (wortmannin, LY 294002, PI 103 and rapamycin)
Many cancers are caused by activating mutations in PI3Kα or inhibitory mutations in PTEN (phosphatase and tensin homologue deleted on chromosome 10), the phosphatase that reconverts PtdIns(3,4,5)P3 (the product of the PI3K reaction) into PtdIns(4,5)P2. For this reason, the development of potent and specific inhibitors of Class 1 PI3Ks has recently become of great interest for the development of novel anti-cancer drugs.
The fungal metabolite wortmannin was originally known as a potent inhibitor of the neutrophil respiratory burst and was shown subsequently to inhibit smMLCK [58]. However, it later became clear that it was a far more potent inhibitor of Class 1 and Class 2 PI3Ks than of MLCK, and it completely suppresses their activities when added to the cell culture medium at only 50–100 nM. More recently, wortmannin was also found to inhibit PLK1 [59]. We therefore re-examined its specificity against our extended panel. These studies confirmed that wortmannin inhibited smMLCK and PLK1 in our assays in the micromolar range (Tables 4 and 5), but no other protein kinases in the panel were inhibited significantly. At micromolar concentrations, wortmannin is also reported to inhibit a PI4K and mTOR (mammalian target of rapamycin), another member of the PI 3K superfamily.
Table 4. Specificities of the PI3K inhibitors TORC1, PDK1 and PKB/AKT.
The concentrations of compounds used in the assays are indicated below each molecule and the results are presented as the percentage activity remaining in the presence of inhibitors compared with control incubations with inhibitor omitted (averages of duplicate determinations). Each experiment was repeated two or three times with similar results. Further details of the assays are given in the Materials and methods section. The effect of the inhibitor Akt-I-1,2 was tested on full-length PKBα as well as the catalytic domain of PKBα for the reasons discussed in the text. Important values are highlighted in boldface type. ND, not determined.
| Percentage activity remaining | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kinase | Wortmannin (1 μM) | LY 294002 (10 μM) | PI-103 (1 μM) | Rapamycin (1 μM) | BX 795 (0.01 μM) | BX 795 (0.1 μM) | BX 320 (1 μM) | BX 320 (10 μM) | A-443654 (0.01 μM) | A-443654 (0.1 μM) | Akt-I-1,2 (1 μM) | Akt-I-1,2 (10 μM) |
| MKK1 | 63±0 | 86±1 | 68±5 | 93±4 | 50±4 | 33±3 | 60±7 | 30±1 | 84±2 | 67±6 | 91±8 | 76±10 |
| ERK1 | 106±5 | 85±7 | 88±6 | 100±2 | 82±9 | 66±10 | 81±9 | 72±3 | 77±8 | 59±3 | 93±8 | 100±6 |
| ERK2 | 90±7 | 88±11 | 96±4 | 94±4 | 88±4 | 71±2 | 84±1 | 73±9 | 78±6 | 64±2 | 99±12 | 76±14 |
| JNK1 | 84±2 | 92±5 | 97±3 | 90±8 | 104±7 | 85±6 | 107±8 | 79±4 | 89±7 | 90±3 | 82±12 | 70±10 |
| JNK2 | 83±0 | 97±2 | 93±4 | 81±1 | 101±12 | 70±4 | 95±4 | 65±7 | 94±1 | 78±9 | 105±3 | 81±12 |
| JNK3 | 97±0 | 86±2 | 90±6 | 81±0 | 93±4 | 93±3 | 97±6 | 102±3 | 81±9 | 86±8 | 88±5 | 80±7 |
| p38α MAPK | 117±9 | 101±10 | 91±5 | 75±4 | 86±2 | 87±1 | 107±6 | 96±7 | 131±2 | 92±6 | 101±11 | 99±14 |
| P38β MAPK | 97±5 | 100±6 | 85±5 | 84±0 | 94±13 | 71±1 | 73±7 | 46±5 | 114±3 | 105±4 | 108±10 | 114±3 |
| p38γ MAPK | 94±1 | 89±3 | 97±8 | 81±7 | 95±2 | 77±2 | 86±4 | 80±1 | 87±5 | 83±4 | 102±12 | 93±10 |
| p38δ MAPK | 83±5 | 102±14 | 81±4 | 92±5 | 91±6 | 76±5 | 93±2 | 75±10 | 86±1 | 68±9 | 111±11 | 98±12 |
| ERK8 | 98±6 | 70±4 | 74±3 | 97±5 | 40±3 | 7±0 | 57±10 | 19±1 | 61±2 | 28±3 | 97±12 | 71±2 |
| RSK1 | 86±5 | 92±3 | 90±13 | 90±3 | 100±6 | 55±10 | 90±3 | 40±7 | 78±1 | 32±1 | 98±5 | 67±1 |
| RSK2 | 90±2 | 104±5 | 105±10 | 97±5 | 100±9 | 75±3 | 88±3 | 42±6 | 74±3 | 27±3 | 80±1 | 74±1 |
| PDK1 | 91±1 | 89±2 | 90±2 | 85±0 | 30±2 | 5±1 | 34±1 | 8±1 | 88±1 | 79±8 | 96±3 | 95±7 |
| PKBα | 97±6 | 95±3 | 75±2 | 91±4 | 90±14 | 90±11 | 82±15 | 57±1 | 5±2 | 3±2 | 84±10 | 64±8 |
| PKBβ | 90±1 | 81±9 | 77±5 | 85±9 | 113±6 | 97±7 | 104±0 | 80±1 | 17±2 | 2±0 | 92±14 | 87±15 |
| SGK1 | 86±6 | 104±4 | 89±3 | 93±1 | 87±6 | 48±3 | 75±5 | 45±2 | 78±2 | 53±5 | 116±13 | 82±3 |
| S6K1 | 95±1 | 91±3 | 82±8 | 106±8 | 100±3 | 84±9 | 100±8 | 62±6 | 88±3 | 32±9 | 100±1 | 72±6 |
| PKA | 86±1 | 97±0 | 74±6 | 77±1 | 102±1 | 72±6 | 99±1 | 59±3 | 26±1 | 3±1 | 91±6 | 77±6 |
| ROCK 2 | 99±3 | 90±11 | 76±4 | 103±3 | 90±6 | 28±1 | 78±4 | 56±6 | 48±3 | 8±1 | 105±11 | 92±1 |
| PRK2 | 71±2 | 75±7 | 81±9 | 79±1 | 79±5 | 43±9 | 72±2 | 42±0 | 17±2 | 2±2 | 80±15 | 68±10 |
| PKCα | 68±1 | 77±7 | 82±5 | 88±3 | 105±1 | 74±1 | 99±2 | 68±0 | 73±3 | 29±4 | 108±4 | 87±8 |
| PKCζ | 84±0 | 114±5 | 77±3 | 92±6 | 96±12 | 95±2 | 83±6 | 80±8 | 82±3 | 66±0 | 105±5 | 96±9 |
| PKD1 | 92±3 | 106±3 | 92±2 | 92±3 | 84±3 | 38±3 | 86±15 | 46±11 | 63±3 | 23±4 | 76±2 | 82±15 |
| MSK1 | 85±1 | 104±0 | 97±2 | 86±2 | 88±0 | 84±5 | 92±0 | 77±8 | 19±2 | 9±2 | 81±2 | 93±14 |
| MNK1 | 81±2 | 77±5 | 90±6 | 83±2 | 80±8 | 37±3 | 88±13 | 75±4 | 120±3 | 109±2 | 77±0 | 84±2 |
| MNK2 | 97±0 | 95±4 | 104±12 | 102±3 | 47±4 | 19±2 | 72±6 | 31±2 | 97±2 | 80±5 | 99±15 | 80±3 |
| MAPKAP-K2 | 125±4 | 103±3 | 99±17 | 108±4 | 108±8 | 108±2 | 103±8 | 68±2 | 92±8 | 73±0 | 84±7 | 60±1 |
| MAPKAP-K3 | 98±5 | 119±5 | 88±16 | 104±5 | 103±6 | 102±1 | 106±4 | 77±6 | 91±9 | 96±2 | 86±6 | 71±5 |
| PRAK | 69±1 | 83±6 | 93±6 | 91±3 | 97±6 | 83±2 | 82±15 | 66±10 | 107±3 | 107±4 | 85±3 | 79±6 |
| CaMKKα | 83±1 | 71±4 | 88±7 | 80±8 | 86±13 | 69±6 | 89±14 | 48±4 | 112±2 | 117±1 | 93±5 | 62±3 |
| CaMKKβ | 71±4 | 71±4 | 79±6 | 83±2 | 82±6 | 30±1 | 63±10 | 20±2 | 83±8 | 63±9 | 138±10 | 108±9 |
| CaMK1 | 97±5 | 87±15 | 108±8 | 97±1 | 86±10 | 71±3 | 71±5 | 52±6 | 89±3 | 77±6 | 43±2 | 3±0 |
| SmMLCK | 72±4 | 88±7 | 62±5 | 107±1 | 110±6 | 95±1 | 104±4 | 95±4 | 69±2 | 27±2 | 79±6 | 11±4 |
| PHK | 74±2 | 94±10 | 84±6 | 95±2 | 94±12 | 62±6 | 102±2 | 77±3 | 86±7 | 54±6 | 103±13 | 101±10 |
| CHK1 | 87±7 | 83±12 | 94±6 | 97±6 | 84±5 | 46±3 | 71±5 | 34±1 | 100±3 | 99±2 | 93±11 | 85±2 |
| CHK2 | 90±3 | 84±0 | 79±12 | 92±3 | 91±1 | 46±4 | 100±2 | 45±0 | 94±8 | 95±2 | 91±12 | 83±7 |
| GSK3β | 65±1 | 37±3 | 84±11 | 83±6 | 82±15 | 48±1 | 55±2 | 23±0 | 81±1 | 33±4 | 89±6 | 78±10 |
| CDK2–cyclin A | 95±9 | 81±2 | 95±5 | 92±4 | 57±7 | 10±0 | 91±7 | 35±1 | 82±5 | 31±2 | 97±0 | 97±14 |
| PLK1 | 64±1 | 26±1 | 79±10 | 78±4 | 95±7 | 92±9 | 97±8 | 70±1 | 89±5 | 80±1 | 79±3 | 90±2 |
| Aurora B | 83±1 | 70±4 | 84±2 | 86±5 | 17±1 | 3±1 | 7±3 | 4±0 | 81±3 | 80±2 | 82±9 | 60±4 |
| Aurora C | 69±3 | 84±2 | 104±4 | 97±5 | 27±3 | 9±1 | 8±1 | 7±1 | 100±4 | 92±8 | 75±6 | 66±0 |
| AMPK | 88±3 | 97±7 | 106±16 | 87±2 | 52±1 | 11±0 | 46±4 | 13±3 | 84±8 | 67±3 | 97±10 | 92±10 |
| MARK3 | 85±3 | 79±13 | 100±6 | 88±4 | 12±1 | 3±0 | 36±4 | 8±0 | 100±3 | 83±1 | 84±12 | 95±12 |
| BRSK2 | 80±4 | 104±4 | 97±1 | 90±4 | 92±4 | 45±3 | 72±3 | 23±4 | 88±9 | 79±1 | 96±8 | 93±13 |
| MELK | 61±2 | 86±9 | 79±9 | 74±5 | 95±4 | 32±3 | 57±10 | 11±1 | 96±2 | 63±2 | 80±2 | 68±0 |
| CK1δ | 94±1 | 57±3 | 87±1 | 88±1 | 79±5 | 91±1 | 89±6 | 96±6 | 95±3 | 74±3 | 105±13 | 111±3 |
| CK2 | 96±7 | 23±5 | 83±6 | 92±9 | 93±11 | 93±0 | 105±10 | 95±7 | 96±0 | 90±8 | 88±13 | 91±14 |
| DYRK1A | 88±1 | 76±10 | 87±5 | 91±4 | 94±7 | 83±3 | 87±4 | 64±8 | 10±2 | 7±3 | 94±15 | 94±12 |
| DYRK2 | 98±1 | 94±3 | 81±7 | 84±9 | 105±10 | 76±6 | 93±0 | 71±0 | 79±5 | 26±1 | 113±8 | 118±1 |
| DYRK3 | 84±1 | 78±2 | 79±8 | 88±1 | 85±9 | 65±15 | 86±9 | 44±1 | 41±3 | 8±1 | 86±4 | 75±2 |
| NEK2a | 81±1 | 91±8 | 114±2 | 82±5 | 92±7 | 95±1 | 95±4 | 61±4 | 101±5 | 95±2 | 91±4 | 100±4 |
| NEK6 | 113±7 | 103±5 | 111±7 | 95±3 | 96±3 | 98±3 | 117±14 | 92±2 | 95±2 | 95±4 | 88±14 | 75±4 |
| NEK7 | 100±7 | 86±1 | 106±2 | 102±5 | 111±8 | 99±2 | 116±14 | 84±11 | 86±8 | 84±1 | 91±7 | 85±10 |
| IKKβ | 96±4 | 68±5 | 95±5 | 82±3 | 108±2 | 93±3 | 96±10 | 69±2 | 93±8 | 89±2 | 98±15 | 90±3 |
| PIM1 | 87±3 | 8±2 | 77±3 | 91±3 | 103±7 | 103±1 | 86±2 | 64±4 | 57±8 | 15±2 | 80±1 | 57±6 |
| PIM2 | 83±4 | 65±2 | 106±7 | 117±0 | 108±3 | 99±1 | 104±10 | 72±2 | 59±0 | 15±2 | 88±3 | 89±3 |
| PIM3 | 86±1 | 13±3 | 79±2 | 88±1 | 96±7 | 63±3 | 97±2 | 54±4 | 54±6 | 13±1 | 84±10 | 60±6 |
| SRPK1 | 73±3 | 101±5 | 82±8 | 84±1 | 99±11 | 86±15 | 79±8 | 70±2 | 92±8 | 91±2 | 84±10 | 84±10 |
| MST2 | 96±9 | 85±1 | 91±6 | 90±5 | 96±2 | 92±11 | 97±4 | 70±6 | 76±4 | 29±0 | 96±11 | 78±2 |
| EF2K | 97±4 | 83±3 | 110±13 | 94±0 | 98±5 | 93±7 | 110±7 | 84±4 | 93±7 | 94±8 | 93±4 | 40±7 |
| HIPK2 | 94±7 | 38±2 | 67±0 | 93±7 | 95±11 | 101±8 | 97±9 | 75±3 | 73±2 | 24±3 | 102±6 | 85±3 |
| HIPK3 | 75±1 | 77±2 | 75±6 | 108±4 | 95±1 | 94±1 | 102±7 | 101±5 | 90±0 | 71±2 | 77±2 | 66±3 |
| PAK4 | 90±2 | 67±5 | 98±5 | 81±1 | 91±15 | 71±4 | 79±7 | 26±1 | 99±3 | 90±6 | 92±4 | 49±5 |
| PAK5 | 72±1 | 92±5 | 91±8 | 89±8 | 97±3 | 81±0 | 93±1 | 41±8 | 104±2 | 96±1 | 96±1 | 61±4 |
| PAK6 | 86±1 | 100±0 | 98±7 | 89±1 | 92±3 | 85±3 | 89±2 | 61±1 | 87±2 | 87±3 | 116±7 | 92±2 |
| Src | 83±1 | 106±11 | 90±8 | 86±1 | 90±4 | 77±2 | 85±1 | 40±2 | 91±0 | 86±5 | 105±10 | 118±1 |
| Lck | 73±4 | 97±2 | 89±3 | 79±7 | 97±4 | 80±11 | 76±4 | 41±2 | 97±8 | 97±3 | 84±2 | 65±6 |
| CSK | 73±9 | 104±3 | 82±1 | 83±1 | 114±12 | 101±7 | 93±15 | 67±11 | 99±2 | 93±7 | 87±9 | 103±15 |
| IKKϵ | ND | ND | ND | ND | 21±5 | 0±6 | 37±4 | 4±9 | ND | ND | 87±3 | 67±6 |
| TBK1 | ND | ND | ND | ND | 7±0 | 1±0 | 32±2 | 5±1 | ND | ND | 101±6 | 107±10 |
| PKBα-full length | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 22±1 | 8±1 |
LY 294002 is another commonly used, but less potent, inhibitor of PI3Ks, which inhibits Class 1 PI3Ks at 10–50 μM in cell-based assays. It has been the inhibitor of choice when cells are incubated for prolonged periods, because wortmannin is unstable in aqueous solution. However, LY 294002 is also reported to inhibit other kinases, such as TORC1, CK2 [1] and PLK1 [59] at concentrations similar to those that inhibit PI3Ks [1]. Using our extended panel, we now find that LY 294002 also inhibits PIM1, PIM3, HIPK2 and GSK3 (Tables 2 and 4), again at concentrations similar to those that inhibit Class 1 PI3Ks. Immobilized LY 294002 was recently shown to bind GSK3 and a number of other ATP-binding proteins that are not protein kinases [60].
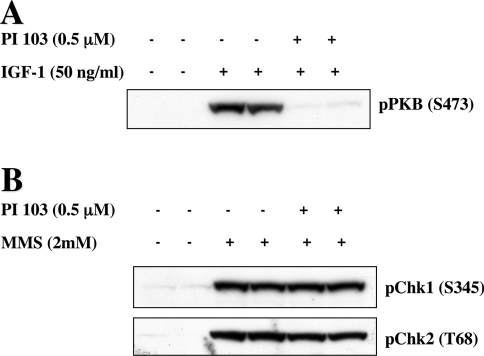
In cell-based assays PI-103 [61] blocks Class 1 PI3Ks completely at only 0.5 μM, as judged by suppression of the IGF-1-stimulated activation of PKB in HEK-293 cells (Figure 2A). However, it inhibited relatively few of the 70 protein kinases in our panel and then by no more than 30–40%, even when assayed in vitro at 1 μM and at low ATP concentrations (Table 4). Moreover, PI 103 at this concentration did not affect two other members of the PI3K superfamily, the protein kinases ATM (ataxia telangiectasia mutated) and ATR (ATM and Rad3-related), as judged by its failure to suppress the phosphorylation (activation) of their substrates, the protein kinases CHK1 and CHK2, in cell-based assays (Figure 2B). However, in another recent study, PI 103 was shown to inhibit TORC1 with similar potency to Class 1 PI3Ks [62].
Figure 2. Effect of PI 103 on the activities of PI3K superfamily members in HEK-293 cells.
(A) HEK-293 (human embryonic kidney-293) cells were incubated for 1 h with (+) or without (−) 0.5 μM PI 103, then stimulated for 30 min with IGF-1 (50 ng/ml) and lysed. Cell extracts (60 μg of protein) were denatured in SDS, subjected to SDS/PAGE and, after transfer to PVDF membranes, immunoblotted with a phosphospecific antibody that recognizes PKB phosphorylated at Ser473 [pPKB (S473)]. (B) HEK-293 cells were incubated for 1 h with (+) or without (−) 0.5 μM PI 103, then treated for 2 h with the DNA-alkylating agent MMS (2 mM) and lysed. Cell extracts (60 μg of protein) were denatured in SDS, subjected to SDS/PAGE and, after transfer to PVDF membranes, immunoblotted with phosphospecific antibodies that recognize CHK1 phosphorylated at Ser345 [pChk1 (S345)]and CHK2 phosphorylated at Thr68 [pChk2 (T68)].
Rapamycin is a naturally occurring compound produced by the soil bacterium Streptomyces hygroscopicus, which originates from Easter Island (Rapa Nui is the native name for Easter Island). It was first purified over 35 years ago as an antifungal agent, but was originally discarded because of its undesirable immunosuppressive side effects. Its potential an as immunosuppressive drug was only explored many years later, and it was finally approved as an immunosuppressant in 1999. It is used most frequently to prevent tissue rejection after kidney and pancreatic islet transplantation. The anticancer properties of rapamycin were also noticed in the mid-1970s, and a modified form of rapamycin has recently been approved for clinical use. Rapamycin exerts its effects on cells by binding to FKBP (FK506 binding protein), and the molecular target for the rapamycin–FKBP complex was identified as TORC1 [63]. The unusual mechanism of action of rapamycin may explain why it does not inhibit any protein kinase in our extended panel (Table 4) or any other protein kinase that has been tested, even at a concentration of 1 μM, which is 10–20-fold higher than that required to inhibit TORC1 activity completely in cell-based assays.
In summary, while wortmannin continues to be very useful as an inhibitor of PI3Ks in cell-based assays, we recommend that the use of LY 294002 be discontinued and that it be replaced by PI-103. Rapamycin is an exquisitely specific inhibitor of TORC1 and should be used in parallel to check whether any of the observed effects of PI-103 result from the inhibition of TORC1, rather than PI3Ks.
PDK1 inhibitors BX 795 and BX 320
PDK1 catalyses the activation of PKB isoforms, a reaction that requires the presence of PtdIns(3,4,5)P3, the product of the PI3K-catalysed reaction. Mice expressing 15% of the normal level of PDK1 are strikingly protected against the formation of multiple tumours that occur in animals carrying only one copy of the PTEN gene [64]. For this reason, PDK1 has become an attractive target for an anticancer drug [65]. BX 795 and BX 320 have been described as potent and specific inhibitors of PDK1 [66] and are beginning to be used to block its activity in cells. In the present study we found that BX 795 was not only a potent inhibitor of PDK1, but also inhibited ERK8, MNK2, Aurora B, Aurora C, MARK3 and IKKϵ with similar potency. TBK1 was inhibited even more potently than PDK1 (Table 4). The IC50 values for inhibition of these protein kinases in our assays were: PDK1 (17 nM), Aurora B (11 nM), IKKϵ (9.5 nM) and TBK1 (2.3 nM). The specificity of BX 320 was similar to BX 795, although it was a much less potent inhibitor.
Interestingly, Aurora kinase (see below) and TBK1 [67,68], like PDK1, are also attractive targets for the development of anticancer drugs. TBK1 is activated in response to hypoxia [67] and controls the production of angiogenic factors such as VEGF (vascular endothelial growth factor) and IL-8 (interleukin-8). Moreover, its levels are elevated in malignant colon and breast-cancer cells. TBK1 is also reported to be activated by the RalB–Sec5 effector complex, restricting the initiation of apoptotic programmes and so aiding tumour-cell survival [68]. BX 795 and other compounds that are potent inhibitors of these three protein kinases might therefore be particularly effective as anticancer agents.
The present study indicates that BX 320 and BX 795 are not specific inhibitors of PDK1, but might be useful for assessing the physiological roles of TBK1 and the closely related IKKϵ, as they are the most potent inhibitors of these two protein kinases to be described thus far.
PKB inhibitors A-443654 and Akt-I-1,2
PKB (also called Akt), a protein kinase that is activated by PDK1 in vivo, has also attracted considerable interest as an anticancer target. A-443654 has been described as a specific inhibitor of PKB [69] and is being used to ascribe particular functions to this protein kinase. In the present study we confirmed that this compound was indeed a very potent inhibitor of PKB, but found that it also inhibits some other members of the AGC subfamily of protein kinases (comprising PKA, PKG and PKC, but also including PKB/Akt, S6K1, RSK1 and PDK1) with slightly lower potency, such as PKA, PRK2 and MSK1, and it also inhibited DYRK1A. Several other protein kinases were inhibited to a lesser extent (Table 4). These analyses show that A-443654 is not a selective PKB inhibitor and should be used with considerable caution.
In contrast with A-443654, Akt-I-1,2 is a highly selective non-competitive inhibitor of PKB in vitro [70]. At a concentration of 1 μM, it inhibits full-length PKBα/AKT1 or CaMK1 by 80%, but no other protein kinase in the panel, including the catalytic domains of PKBα and PKBβ, was inhibited significantly at this concentration (Table 4). This is because inhibition by Akt-I-1,2 requires the presence of the PH (pleckstrin homology) domain. Importantly, Akt-1-I/2 prevents the conformational change, triggered by the binding of PtdIns(3,4,5)P3 to the PH domains of PKB isoforms, that allows PDK1 and TORC2 to phosphorylate and activate PKB. For this reason, Akt-I-1,2 is a potent inhibitor of the activation of PKB rather than of the active PKB itself, and prevents the insulin-induced activation of PKB/Akt when added to cells at 1 μM [71].
In summary, we recommend the use of Akt-I-1,2 to inhibit PKB activation in cells.
Inhibitors of CK1 (D4476, CK1-7 and IC 261)
CK1 isoforms play multiple roles in cell regulation. We have previously reported that the compound D4476 synthesized during a programme to develop inhibitors of ALK5 (activin-receptor-like kinase 5) was a relatively selective inhibitor of CK1 and more potent than the other known CK1 inhibitors CK1-7 and IC261 against 30 protein kinases [72]. Here we extended these studies to the larger panel (Tables 5 and 6). The results confirmed that D4476 is a rather selective inhibitor of CK1. D4476 inhibited CK1δ 20–30-fold more potently than PKD1 or p38α MAPK, and no other protein kinases in the panel were inhibited to a significant extent. CK1-7 and IC261 were 5–10-fold less potent inhibitors of CK1 and also inhibited several other protein kinases, including PIM1 and PIM3 (CK1-7 and IC261), ERK8, MNK1, AMPK, SGK1 (CK1-7) (Table 6). We recommend the use of D4476 to inhibit CK1 isoforms in cell-based assays. A method for preventing its precipitation in aqueous solution has been described [72].
Table 6. Specificities of compounds identified as CK1 and DYRK inhibitors.
The concentrations of compounds used in the assays are indicated below each molecule and the results are presented as the percentage activity remaining in the presence of inhibitor as compared with control incubations with inhibitor omitted (averages of duplicate determinations). Each experiment was repeated two or three times with similar results. Further details of the assays are given in the Materials and methods section. Important values are highlighted in boldface type.
| Percentage activity remaining | |||||||
|---|---|---|---|---|---|---|---|
| Kinase | D4476 (10 μM) | CK1-7 (25 μM) | IC261 (25 μM) | Harmine (1 μM) | Harmane (1 μM) | Harmalol (1 μM) | Harmaline (1 μM) |
| MKK1 | 93±2 | 76±2 | 86±2 | 73±4 | 90±6 | 86±1 | 95±3 |
| ERK1 | 102±4 | 102±4 | 91±6 | 95±5 | 107±1 | 107±1 | 108±4 |
| ERK2 | 85±2 | 100±2 | 93±4 | 95±5 | 95±6 | 95±6 | 87±0 |
| JNK1 | 86±5 | 91±1 | 94±2 | 89±3 | 92±4 | 102±4 | 96±3 |
| JNK2 | 77±7 | 87±9 | 101±3 | 91±5 | 84±6 | 99±6 | 86±7 |
| JNK3 | 82±0 | 103±0 | 93±4 | 91±0 | 89±2 | 113±0 | 89±0 |
| p38α MAPK | 38±1 | 103±5 | 92±4 | 100±7 | 74±1 | 96±2 | 88±4 |
| p38β MAPK | 55±2 | 101±9 | 97±7 | 103±4 | 90±7 | 95±4 | 92±1 |
| p38γ MAPK | 87±1 | 102±3 | 93±4 | 80±3 | 97±9 | 104±2 | 120±6 |
| p38δ MAPK | 89±2 | 80±8 | 91±4 | 88±1 | 90±1 | 89±4 | 92±5 |
| ERK8 | 80±4 | 29±6 | 68±2 | 80±1 | 70±0 | 62±3 | 44±4 |
| RSK1 | 81±1 | 48±3 | 87±3 | 105±6 | 98±1 | 100±9 | 110±5 |
| RSK2 | 82±1 | 63±4 | 89±1 | 104±3 | 94±1 | 83±8 | 112±1 |
| PDK1 | 85±1 | 83±3 | 89±5 | 87±1 | 90±7 | 93±6 | 86±9 |
| PKBα | 107±9 | 54±1 | 94±5 | 101±7 | 104±3 | 89±2 | 95±0 |
| PKBβ | 88±1 | 89±4 | 90±3 | 90±1 | 91±8 | 81±1 | 105±1 |
| SGK1 | 73±2 | 25±2 | 79±4 | 83±1 | 86±4 | 86±3 | 85±7 |
| S6K1 | 76±5 | 49±5 | 84±2 | 93±3 | 94±3 | 94±8 | 93±1 |
| PKA | 85±6 | 65±7 | 82±3 | 80±9 | 95±2 | 90±4 | 89±1 |
| ROCK 2 | 105±1 | 65±6 | 78±0 | 80±4 | 104±1 | 91±8 | 79±6 |
| PRK2 | 107±4 | 47±3 | 87±5 | 86±4 | 85±6 | 86±3 | 85±1 |
| PKCα | 94±5 | 86±3 | 90±2 | 82±4 | 90±5 | 92±1 | 90±9 |
| PKCζ | 81±1 | 52±1 | 80±4 | 83±1 | 89±3 | 88±0 | 84±3 |
| PKD1 | 43±4 | 60±3 | 62±3 | 80±9 | 88±1 | 111±3 | 108±1 |
| MSK1 | 90±7 | 42±2 | 88±1 | 83±5 | 91±11 | 96±1 | 94±1 |
| MNK1 | 81±1 | 26±0 | 79±3 | 82±4 | 82±1 | 97±1 | 80±1 |
| MNK2 | 88±4 | 76±0 | 97±5 | 80±4 | 112±4 | 106±5 | 92±2 |
| MAPKAP-K2 | 99±4 | 85±3 | 88±4 | 110±4 | 109±3 | 88±1 | 89±4 |
| MAPKAP-K3 | 98±3 | 86±7 | 94±3 | 98±6 | 104±6 | 99±9 | 97±5 |
| PRAK | 77±2 | 81±4 | 82±5 | 90±2 | 84±8 | 85±3 | 74±1 |
| CaMKKα | 89±1 | 86±4 | 67±4 | 95±4 | 86±6 | 103±1 | 92±1 |
| CaMKKβ | 100±2 | 86±3 | 86±2 | 81±1 | 93±5 | 90±1 | 89±7 |
| CaMK1 | 88±2 | 82±3 | 55±5 | 94±4 | 91±4 | 101±3 | 103±0 |
| SmMLCK | 81±5 | 84±7 | 70±2 | 87±7 | 100±4 | 107±9 | 112±2 |
| PHK | 89±4 | 72±5 | 86±1 | 85±5 | 102±3 | 87±3 | 84±7 |
| CHK1 | 84±8 | 86±2 | 99±2 | 99±9 | 99±4 | 84±5 | 94±5 |
| CHK2 | 92±5 | 64±1 | 76±4 | 100±0 | 92±3 | 122±1 | 117±5 |
| GSK3β | 88±5 | 75±4 | 74±6 | 87±0 | 108±1 | 88±4 | 100±9 |
| CDK2-Cyclin A | 113±8 | 62±4 | 83±4 | 86±4 | 103±6 | 101±1 | 100±7 |
| PLK1 | 90±9 | 81±4 | 95±1 | 97±1 | 81±2 | 83±1 | 80±1 |
| Aurora B | 88±300 | 78±0 | 75±3 | 81±2 | 106±3 | 90±8 | 90±4 |
| Aurora C | 81±4 | 95±3 | 94±3 | 83±1 | 87±3 | 91±6 | 99±8 |
| AMPK | 98±0 | 29±5 | 71±2 | 72±1 | 112±13 | 110±1 | 106±1 |
| MARK3 | 82±4 | 78±5 | 88±5 | 83±3 | 128±4 | 131±9 | 110±5 |
| BRSK2 | 90±5 | 42±1 | 88±4 | 83±3 | 101±1 | 98±4 | 103±1 |
| MELK | 83±1 | 63±4 | 75±8 | 80±2 | 82±3 | 77±0 | 95±0 |
| CK1δ | 4±0 | 12±1 | 9±2 | 60±4 | 99±6 | 94±9 | 65±5 |
| CK2 | 83±5 | 65±1 | 97±2 | 68±5 | 85±8 | 91±1 | 87±1 |
| DYRK1A | 86±7 | 39±6 | 73±1 | 4±2 | 44±4 | 67±6 | 29±4 |
| DYRK2 | 93±3 | 76±3 | 99±6 | 15±0 | 83±5 | 86±4 | 73±1 |
| DYRK3 | 76±1 | 46±1 | 89±3 | 6±0 | 72±4 | 71±2 | 49±5 |
| NEK2a | 89±7 | 85±4 | 75±6 | 94±1 | 79±0 | 82±4 | 96±5 |
| NEK6 | 79±1 | 90±2 | 95±5 | 87±1 | 90±1 | 93±1 | 94±1 |
| NEK7 | 100±6 | 102±3 | 103±4 | 108±1 | 105±1 | 109±8 | 107±9 |
| IKKβ | 63±3 | 87±2 | 90±5 | 73±9 | 76±8 | 74±1 | 94±5 |
| PIM1 | 93±8 | 28±2 | 43±1 | 86±1 | 94±1 | 92±1 | 104±7 |
| PIM2 | 90±4 | 55±6 | 75±1 | 88±7 | 85±1 | 101±1 | 117±1 |
| PIM3 | 82±4 | 28±1 | 41±1 | 64±1 | 80±4 | 71±1 | 90±6 |
| SRPK1 | 84±5 | 86±2 | 95±1 | 86±7 | 81±3 | 88±8 | 89±6 |
| MST2 | 96±2 | 91±0 | 70±1 | 85±7 | 81±1 | 90±3 | 101±1 |
| EF2K | 100±1 | 94±6 | 100±4 | 104±8 | 101±3 | 102±6 | 107±2 |
| HIPK2 | 80±4 | 98±7 | 96±6 | 108±7 | 104±5 | 101±1 | 91±1 |
| HIPK3 | 96±3 | 86±7 | 95±3 | 87±8 | 104±0 | 93±8 | 97±1 |
| PAK4 | 105±1 | 88±3 | 84±2 | 96±3 | 103±5 | 105±6 | 90±1 |
| PAK5 | 100±1 | 86±7 | 84±1 | 92±5 | 97±8 | 95±4 | 84±7 |
| PAK6 | 103±5 | 78±3 | 76±0 | 83±5 | 105±3 | 92±1 | 106±2 |
| Src | 82±0 | 92±1 | 70±4 | 88±9 | 83±2 | 85±5 | 94±6 |
| Lck | 82±5 | 83±6 | 49±4 | 81±2 | 93±1 | 93±1 | 81±1 |
| CSK | 81±6 | 79±5 | 81±4 | 86±3 | 81±5 | 84±7 | 82±7 |
Identification of harmine as a specific inhibitor of DYRK isoforms
Healers in the Amazon region have been using harmine as a psychoactive compound in a brew known as ‘ayahuasca’ for thousands of years. A serotonin antagonist and reversible short-term inhibitor of monoamine oxidase, it was first used to treat Parkinsonism in 1928, where it was said to brighten the mental status of the patients. However, it is also of interest as an anti-cancer agent and, in this connection, was reported to inhibit CDKs in the micromolar range [73]. These findings led us to examine its specificity against our panel of protein kinases, which revealed that harmine was an extremely potent and specific inhibitor of the DYRK family of protein kinases (Table 6). It inhibited DYRK1A in the nanomolar range, the DYRK2 and DYRK3 isoforms being inhibited about 10-fold less potently. In our experiments, harmine did not inhibit CDK2 significantly, but did inhibit the three PIM isoforms and CK1 in the micromolar range (Tables 5 and 6).
Down's syndrome, resulting from the presence of an extra copy of chromosome 21, is the most common genetic disorder in humans, with a frequency of 1 in 800 live births. The Down's-syndrome child begins life with an IQ close to that of a normal child, but these parameters gradually deteriorate until, at age 13, they display an average IQ of 50. Interestingly, the gene encoding DYRK1A is located within the Down's syndrome critical region of chromosome 21. DYRK1A is expressed at elevated levels in human Down's-syndrome foetal tissues, and mice that overexpress this kinase have defects in neural development [74]. A recent report suggested that the pathological effects of high DYRK1A activity may result from the hyperphosphorylation and reduced activity of the transcription factor NFATc (nuclear factor of activated T-cells) [75]. The finding that harmine is a potent and specific inhibitor of DYRK1A raises the possibility of preventing mental retardation in Down's-syndrome patients through the use of drugs, such as harmine or a derivative of this compound, that inhibit this protein kinase.
Very recently, harmine was identified as an anti-diabetic, cell-type-specific regulator of PPARγ (peroxisome-proliferator-activated receptor γ) expression and, when administered to diabetic mice, it mimicked the effect of PPARγ ligands on adipocyte gene expression and sensitivity to insulin [76]. It will clearly be of great interest to find out whether the anti-diabetic effects of harmine are explained by its ability to inhibit one or more DYRK isoforms.
The potent inhibition of DYRK1A by harmine was unexpected, given its rather low molecular mass (212 Da), and understanding how this drug interacts with DYRK1A will be of considerable interest. The related compounds, harmalol, harmaline and harmane were also relatively specific, but much weaker, inhibitors of DYRK isoforms (Table 6). However, currently there is no information at to whether harmine can suppress the activity of DYRK1A in cells.
CDK inhibitors roscovitine and purvalanol A
The olomoucine derivatives roscovitine [77] and purvalanol [78] were identified as CDK inhibitors a number of years ago. Purvalanol was found to inhibit several protein kinases in our panel, such as PAK4, PAK5, MELK, Src (Tables 5 and 7) and Yes (results not shown), although not as potently as CDK2. Roscovitine inhibited ERK8, but was only a weak inhibitor of other protein kinases (Tables 5 and 7). Roscovitine and purvalanol are known to inhibit other CDKs with similar potency to CDK2, including CDK1, CDK5 and CDK7 [77,78], whereas roscovitine also inhibits pyridoxal kinase [79]. These findings support the continued use of these two compounds as pan-CDK inhibitors.
Table 7. Specificities of compounds developed as inhibitors of CDK, Aurora kinases, CaMKK, AMPK, IKK, JNK and MNK.
The concentrations of compounds used in the assays are indicated below each molecule and the results are presented as the percentage activity remaining in the presence of inhibitors as compared with control incubations with inhibitor omitted (averages of duplicate determinations). Each experiment was repeated two or three times with similar results. Further details of the assays are given in the Materials and methods section. Important values are highlighted in boldface type. ND, not determined.
| Percentage activity remaining | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kinase | Roscovitine (1 μM) | Purvalanol (0.1 μM) | VX 680 (1 μM) | SU 6668 (1 μM) | STO609 (1 μM) | Compound C (1 μM) | PS 1145 (10 μM) | BMS 345541 (10 μM) | SC 514 (10 μM) | SP 600125 (1 μM) | AS 601245 (1 μM) | AS 601245 10μM | CGP 57380 (1 μM) |
| MKK1 | 81±6 | 104±7 | 48±1 | 31±1 | 88±5 | 97±3 | 83±1 | 66±4 | 83±8 | 58±7 | 73±6 | 51±6 | 56±1 |
| ERK1 | 82±2 | 95±4 | 96±5 | 85±3 | 98±9 | 110±6 | 106±8 | 87±6 | 88±6 | 89±4 | 104±5 | 97±3 | 78±4 |
| ERK2 | 83±2 | 79±5 | 96±1 | 92±3 | 92±3 | 87±7 | 84±3 | 92±6 | 96±9 | 86±3 | 91±9 | 68±4 | 82±3 |
| JNK1 | 89±5 | 100±4 | 100±2 | 99±14 | 84±4 | 97±6 | 86±4 | 101±0 | 89±4 | 23±1 | 82±9 | 43±4 | 102±7 |
| JNK2 | 88±6 | 96±3 | 102±8 | 99±3 | 69±3 | 92±0 | 93±1 | 98±1 | 93±7 | 24±3 | 73±5 | 29±3 | 97±4 |
| JNK3 | 87±2 | 100±4 | 96±1 | 90±10 | 84±0 | 82±5 | 92±0 | 95±0 | 94±9 | 47±0 | 72±5 | 40±4 | 96±0 |
| p38α MAPK | 96±4 | 95±5 | 104±1 | 95±6 | 93±6 | 93±10 | 89±4 | 101±3 | 102±9 | 110±2 | 72±8 | 63±1 | 82±6 |
| P38β MAPK | 94±3 | 90±2 | 91±5 | 78±5 | 91±1 | 89±7 | 84±6 | 101±2 | 99±8 | 106±2 | 96±5 | 87±8 | 102±1 |
| p38γ MAPK | 99±2 | 94±8 | 98±5 | 84±2 | 103±2 | 82±8 | 98±4 | 96±6 | 79±8 | 86±1 | 78±8 | 46±4 | 102±5 |
| p38δ MAPK | 92±7 | 98±4 | 90±2 | 88±9 | 86±6 | 88±3 | 78±7 | 91±3 | 87±1 | 82±2 | 68±10 | 28±2 | 101±5 |
| ERK8 | 40±5 | 70±6 | 93±2 | 51±1 | 19±5 | 9±1 | 42±6 | 56±2 | 47±2 | 43±1 | 48±3 | 12±2 | 86±6 |
| RSK1 | 97±2 | 74±2 | 31±5 | 55±7 | 85±4 | 62±3 | 66±1 | 91±8 | 97±5 | 55±0 | 71±6 | 40±1 | 51±2 |
| RSK2 | 71±3 | 71±3 | 57±6 | 50±3 | 105±1 | 66±7 | 82±4 | 92±4 | 90±6 | 70±1 | 89±8 | 52±6 | 83±1 |
| PDK1 | 80±8 | 79±5 | 91±4 | 73±7 | 83±1 | 94±8 | 87±3 | 98±6 | 93±11 | 56±6 | 102±6 | 92±9 | 74±3 |
| PKBα | 100±3 | 95±6 | 103±6 | 93±11 | 92±4 | 72±7 | 90±1 | 88±5 | 81±7 | 97±1 | 78±4 | 32±3 | 96±4 |
| PKBβ | 83±4 | 92±3 | 102±3 | 95±1 | 107±1 | 65±14 | 101±9 | 93±1 | 103±8 | 92±1 | 95±7 | 51±5 | 90±5 |
| SGK1 | 75±2 | 82±1 | 80±7 | 80±3 | 96±6 | 80±5 | 75±0 | 91±4 | 105±6 | 28±0 | 92±3 | 69±5 | 57±2 |
| S6K1 | 90±5 | 87±3 | 84±2 | 47±4 | 74±1 | 59±4 | 86±1 | 83±3 | 84±9 | 85±1 | 96±4 | 65±8 | 98±4 |
| PKA | 73±1 | 79±5 | 60±6 | 102±10 | 93±1 | 85±9 | 86±8 | 79±3 | 85±2 | 80±6 | 112±8 | 77±8 | 71±5 |
| ROCK 2 | 90±4 | 93±6 | 86±1 | 68±5 | 99±1 | 67±6 | 69±6 | 80±5 | 59±3 | 92±2 | 80±3 | 32±7 | 97±5 |
| PRK2 | 89±2 | 81±3 | 61±7 | 73±3 | 73±1 | 37±8 | 71±7 | 84±7 | 35±1 | 70±5 | 88±12 | 58±8 | 90±5 |
| PKCα | 89±2 | 77±1 | 93±3 | 87±2 | 77±4 | 92±11 | 77±2 | 88±3 | 67±6 | 68±5 | 89±6 | 80±7 | 90±6 |
| PKCζ | 82±6 | 83±7 | 76±6 | 97±1 | 84±1 | 94±7 | 78±6 | 84±4 | 75±10 | 106±2 | 86±4 | 85±12 | 88±5 |
| PKD1 | 90±4 | 85±7 | 100±4 | 82±5 | 81±4 | 94±6 | 108±4 | 54±5 | 75±2 | 26±0 | 108±10 | 40±11 | 86±4 |
| MSK1 | 78±1 | 89±0 | 86±1 | 74±13 | 91±1 | 64±4 | 99±1 | 82±2 | 86±8 | 72±9 | 89±12 | 69±5 | 85±4 |
| MNK1 | 90±9 | 92±5 | 88±4 | 83±2 | 10±1 | 19±3 | 44±2 | 91±6 | 72±4 | 67±4 | 68±10 | 50±8 | 35±2 |
| MNK2 | 97±2 | 98±1 | 96±2 | 89±6 | 50±7 | 64±1 | 70±2 | 92±2 | 91±3 | 59±5 | 70±6 | 39±4 | 56±2 |
| MAPKAP-K2 | 97±7 | 106±7 | 83±6 | 104±8 | 95±4 | 80±3 | 83±4 | 100±3 | 97±3 | 102±2 | 85±7 | 54±3 | 118±2 |
| MAPKAP-K3 | 86±5 | 88±1 | 87±1 | 103±7 | 102±1 | 102±5 | 95±9 | 94±3 | 92±3 | 103±1 | 105±8 | 96±5 | 104±4 |
| PRAK | 83±4 | 91±2 | 86±2 | 86±13 | 79±2 | 96±6 | 85±5 | 91±7 | 61±6 | 58±9 | 97±8 | 67±3 | 96±4 |
| CAMKKα | 73±5 | 76±1 | 89±4 | 55±5 | 26±0 | 47±6 | 80±1 | 84±1 | 90±4 | 68±7 | 90±11 | 67±8 | 102±2 |
| CAMKKβ | 71±5 | 60±1 | 86±7 | 61±2 | 8±1 | 65±4 | 69±8 | 80±6 | 87±1 | 41±4 | 91±7 | 41±4 | 90±5 |
| CAMK1 | 104±3 | 99±1 | 86±4 | 76±7 | 92±7 | 85±5 | 73±1 | 77±4 | 87±7 | 73±2 | 101±2 | 106±7 | 95±1 |
| SmMLCK | 84±3 | 86±2 | 102±7 | 61±5 | 81±1 | 52±4 | 88±1 | 87±3 | 80±7 | 34±4 | 90±8 | 57±5 | 99±1 |
| PHK | 93±1 | 85±3 | 58±2 | 48±7 | 84±1 | 6±2 | 85±7 | 88±4 | 103±8 | 31±0 | 85±3 | 62±6 | 61±3 |
| CHK1 | 87±5 | 95±4 | 77±3 | 63±1 | 87±0 | 93±5 | 90±5 | 86±3 | 69±9 | 63±3 | 97±1 | 95±9 | 84±6 |
| CHK2 | 93±4 | 94±7 | 93±4 | 16±0 | 90±1 | 95±3 | 102±8 | 86±7 | 93±12 | 44±5 | 95±4 | 64±5 | 95±2 |
| GSK3β | 76±6 | 79±6 | 78±4 | 83±8 | 43±1 | 81±9 | 46±6 | 82±7 | 76±2 | 81±4 | 2±1 | 1±1 | 87±6 |
| CDK2-Cyclin A | 4±0 | 9±1 | 86±2 | 87±6 | 64±2 | 88±3 | 76±0 | 57±4 | 84±7 | 50±1 | 86±6 | 62±5 | 81±1 |
| PLK1 | 91±1 | 81±3 | 94±6 | 83±1 | 83±2 | 106±2 | 98±6 | 95±7 | 99±7 | 80±3 | 106±7 | 43±6 | 91±2 |
| Aurora B | 94±6 | 89±5 | 7±1 | 3±0 | 60±1 | 40±3 | 70±6 | 70±5 | 50±4 | 15±1 | 84±7 | 57±5 | 64±4 |
| Aurora C | 96±4 | 85±3 | 17±1 | 10±0 | 68±4 | 57±7 | 80±6 | 91±5 | 70±2 | 20±3 | 70±6 | 48±7 | 82±4 |
| AMPK | 91±2 | 89±1 | 80±6 | 52±5 | 18±1 | 27±4 | 88±4 | 86±4 | 77±10 | 44±5 | 82±7 | 70±4 | 101±8 |
| MARK3 | 87±5 | 89±8 | 74±3 | 70±6 | 109±1 | 54±3 | 89±4 | 90±6 | 64±5 | 49±5 | 86±9 | 50±7 | 80±1 |
| BRSK2 | 79±4 | 76±6 | 62±1 | 53±2 | 93±4 | 46±4 | 90±1 | 84±4 | 77±2 | 44±8 | 92±7 | 73±6 | 39±8 |
| MELK | 88±7 | 72±3 | 20±1 | 84±6 | 77±5 | 5±0 | 70±1 | 79±3 | 80±5 | 15±4 | 92±3 | 66±7 | 68±6 |
| CK1δ | 76±1 | 103±5 | 104±5 | 57±6 | 68±0 | 43±4 | 92±1 | 55±5 | 91±12 | 15±0 | 71±7 | 51±6 | 30±3 |
| CK2 | 92±7 | 99±1 | 102±1 | 82±8 | 10±1 | 66±5 | 87±1 | 87±4 | 75±6 | 57±4 | 63±6 | 22±3 | 87±3 |
| DYRK1A | 87±5 | 88±7 | 91±6 | 90±7 | 48±1 | 12±1 | 47±2 | 81±2 | 41±1 | 40±1 | 33±3 | 9±2 | 91±6 |
| DYRK2 | 90±4 | 91±4 | 83±2 | 96±12 | 34±1 | 24±7 | 89±1 | 78±2 | 66±4 | 19±1 | 12±1 | 4±3 | 77±3 |
| DYRK3 | 97±2 | 89±4 | 102±5 | 95±8 | 32±1 | 9±1 | 50±3 | 71±2 | 47±6 | 30±8 | 24±3 | 9±5 | 67±5 |
| NEK2a | 85±6 | 95±1 | 90±2 | 87±2 | 84±1 | 74±12 | 85±6 | 90±7 | 90±5 | 78±1 | 83±10 | 81±6 | 91±2 |
| NEK6 | 92±4 | 91±5 | 86±1 | 98±5 | 105±1 | 79±8 | 83±1 | 92±6 | 93±9 | 102±4 | 84±9 | 63±10 | 93±1 |
| NEK7 | 87±5 | 105±5 | 102±1 | 99±9 | 109±1 | 58±5 | 103±4 | 98±5 | 114±1 | 112±7 | 85±7 | 70±5 | 103±7 |
| IKKβ | 80±3 | 89±9 | 84±5 | 90±8 | 78±4 | 98±2 | 4±0 | 29±0 | 28±1 | 49±3 | 94±6 | 86±3 | 85±3 |
| PIM1 | 83±5 | 93±3 | 88±2 | 90±7 | 22±1 | 80±9 | 13±1 | 84±3 | 40±6 | 39±3 | 8±1 | 5±3 | 83±5 |
| PIM2 | 106±4 | 96±6 | 99±3 | 111±12 | 9±3 | 109±8 | 47±1 | 95±1 | 55±2 | 58±2 | 40±5 | 7±2 | 90±1 |
| PIM3 | 83±9 | 91±0 | 77±6 | 66±7 | 1±1 | 85±9 | 9±1 | 64±2 | 25±4 | 17±0 | 3±1 | 2±0 | 54±2 |
| SRPK1 | 84±4 | 79±0 | 82±6 | 67±2 | 60±9 | 78±4 | 88±2 | 92±5 | 82±3 | 84±5 | 96±7 | 72±1 | 98±6 |
| MST2 | 95±5 | 60±3 | 54±2 | 53±3 | 75±0 | 96±4 | 88±7 | 92±7 | 69±6 | 31±4 | 80±4 | 86±3 | 72±6 |
| EF2K | 85±8 | 95±1 | 93±3 | 95±9 | 107±1 | 114±2 | 100±2 | 99±3 | 113±6 | 118±1 | 110±10 | 90±5 | 90±2 |
| HIPK2 | 84±5 | 99±4 | 93±4 | 68±1 | 35±5 | 26±1 | 67±3 | 83±5 | 47±2 | 35±8 | 29±3 | 7±1 | 94±1 |
| HIPK3 | 90±1 | 101±2 | 94±5 | 95±0 | 68±8 | 63±5 | 89±2 | 96±9 | 77±7 | 56±5 | 61±4 | 18±2 | 106±0 |
| PAK4 | 90±3 | 43±0 | 38±1 | 79±7 | 84±1 | 83±4 | 93±1 | 81±4 | 88±4 | 60±4 | 84±11 | 38±3 | 76±4 |
| PAK5 | 74±3 | 53±3 | 60±3 | 88±6 | 86±1 | 93±6 | 87±2 | 84±2 | 82±3 | 82±1 | 86±11 | 54±3 | 87±1 |
| PAK6 | 80±2 | 67±5 | 69±6 | 87±4 | 97±1 | 92±5 | 101±0 | 77±8 | 89±11 | 72±5 | 102±3 | 89±11 | 79±5 |
| Src | 89±2 | 14±2 | 23±3 | 85±1 | 102±1 | 25±2 | 98±5 | 90±7 | 90±6 | 89±1 | 79±4 | 38±3 | 67±6 |
| Lck | 76±2 | 74±5 | 10±4 | 82±5 | 93±1 | 20±2 | 57±3 | 79±4 | 70±7 | 68±2 | 83±5 | 68±7 | 70±6 |
| CSK | 88±4 | 82±2 | 71±7 | 93±8 | 96±5 | 69±6 | 83±1 | 80±0 | 81±2 | 65±4 | 98±4 | 77±6 | 67±1 |
| IKKϵ | ND | ND | ND | 51±8 | ND | 51±7 | 81±3 | 93±2 | 89±4 | ND | ND | ND | ND |
| TBK1 | ND | ND | ND | 34±4 | ND | 45±8 | 87±1 | 93±2 | 93±1 | ND | ND | ND | ND |
| IKKα | ND | ND | ND | ND | ND | ND | 94±1 | 97±1 | 95±5 | ND | ND | ND | ND |
Aurora kinase inhibitors VX 680 and SU6668
VX 680 was developed as a potent inhibitor of Aurora kinases, which regulate several aspects of the cell division cycle, including microtubule–kinetochore attachments. For this reason Aurora kinases are being targeted for the development of anti-cancer drugs, and some have entered clinical trials. More recently, VX 680 was also found to be a potent inhibitor of the Abl (Abelson) protein tyrosine kinase [80]. We found that VX 680 also inhibited MELK, Src and other protein kinases, such as FGF-R1 and Eph-A2 (results not shown), with less marked inhibition of several other protein kinases such as ERK8, RSK1, RSK2, PAK4 and MST2 (Tables 5 and 7). VX 680 has also been reported to inhibit the protein tyrosine kinase FLT3 (fibromyalgia syndrome-related tyrosine kinase 3), although not as potently as Aurora kinases [81]. VX 680 appears to inactivate Aurora A and Aurora B completely when added to the cell culture medium at 1 μM, as judged by the blockade of TACC3 (transforming acidic coiled coil protein 3; an Aurora A substrate) and histone H3 (an Aurora B substrate) [64].
SU 6668 was developed to inhibit the VEGF receptor and FGFR with the aim of inhibiting tumour growth by suppressing angiogenesis, but it has recently been found to bind to and inhibit several other protein kinases, including Aurora kinases, TBK1 and AMPK [82]. When profiled against our extended panel, we found that SU 6668 inhibited not only these protein kinases, but a number of others. MKK1, CHK2, ERK8, RSK1, RSK2, S6K1, Aurora B and Aurora C were the protein kinases inhibited most potently (Tables 5 and 7). These findings indicate that SU 6668 is insufficiently specific to be useful as a protein kinase inhibitor in cell-based assays.
CaMKK inhibitor STO 609
STO 609 has been identified as an inhibitor of CaMKKα and CaMKKβ, which are ‘upstream’ activators of CaMK 1 and 4. CaMKKβ also activates AMPK in neuronal cells [83] and T-cells [84]. When tested against our extended panel, CaMKKβ was inhibited about 10-fold more potently than CaMKKα. However, STO 609 was also inhibited ERK8, MNK1, CK2, AMPK, PIM2, PIM3, DYRK2, DYRK3 and HIPK2 with similar potency to CaMKKα (Tables 5 and 7). STO 609 suppresses CaMKK activity almost completely when added to cells at 25 μM (D. G. Hardie, personal communication). However, although this compound has been used to implicate CaMKKs in the activation of AMPK, the present study indicates that STO 609 is not a specific inhibitor and results obtained by using it should be interpreted with caution.
An inhibitor of AMPK (Compound C)
This compound has been described as an inhibitor of AMPK and is being used increasingly to inhibit this protein kinase in cell-based assays. In the present study we found that Compound C inhibited AMPK with an IC50 value of 0.1–0.2 μM, but a number of other protein kinases were inhibited with similar or greater potency, including ERK8, MNK1, PHK, MELK, DYRK isoforms, HIPK2, Src, Lck (Table 7) and Yes, FGF-R1 and Eph-A2 (results not shown). Since a concentration of 40 μM in the culture medium is needed to inhibit AMPK completely in cells (D. G. Hardie, personal communication), the use of this compound to identify potential functions of AMPK is not recommended.
Inhibitors of IKKβ (PS1145, BMS 345541 and SC 514)
These compounds have been described and used as inhibitors of the IKKs in many studies. PS 1145 inhibited IKKβ with an IC50 value of 0.25 μM (Table 5). It also inhibited PIM1 and PIM3 with similar potency to IKKβ and several other protein kinases with lower potency, but did not inhibit the other three members of the IKK subfamily (IKKα, IKKϵ or TBK1) significantly (Table 7). BMS 345541 and SC 514 inhibited IKKβ about 10-fold more weakly than PS 1145 and also did not inhibit IKKα, IKKϵ and TBK1 (Tables 5 and 7). BMS 345541 inhibited several other kinases with slightly lower potency than IKKβ, including ERK8, PKD1, CDK2 and CK1, whereas SC514 inhibited PIM3, PIM1, DYRK1A, DYRK3 and Aurora B similarly to IKKβ (Table 7).
When added to the cell culture medium at 50 μM, PS 1145 was reported to suppress the LPS (lipopolysaccharide)-induced phosphorylation and activation of the protein kinase Cot/Tpl2 (cancer Osaka thyroid/tumour progression locus-2) at Thr290 [85], leading to the conclusion that the phosphorylation of this residue was catalysed by IKKβ. However, at a lower concentration (15 μM), no suppression of IL-1 (interleukin 1)-induced phosphorylation of Thr290 was observed, even though IKKβ was still blocked completely, as shown by suppression of the degradation of IκBα (inhibitor of nuclear factor κB). This suggested that Thr290 is phosphorylated by a protein kinase distinct from IKKβ [86], the blockade of Thr290 phosphorylation observed at a higher (50 μM) PS 1145 concentration, presumably resulting from the ‘non-specific’ inhibition of another protein kinase.
These findings suggest that results obtained by using PS 1145 should be interpreted with caution and that the development of more specific inhibitors of IKK isoforms would be extremely useful.
JNK inhibitors SP 600125 and AS 601245
We have reported previously that SP 600125 is not a specific inhibitor of JNK, since it inhibited 13 of the 30 protein kinases tested with similar or greater potency than JNK isoforms [2]. However, despite the availability of this information, many laboratories have continued to use SP 600125 as a JNK inhibitor. Further analysis against our extended panel confirmed the lack of specificity of this compound and identified a number of other protein kinases that are inhibited by SP 600125. Those inhibited as potently or more potently than JNK isoforms, include PKD1, CHK2, Aurora B and C, MELK, CK1, DYRK2, DYRK3 and HIPK3 (Table 7).
AS 601245 has also been reported as a JNK inhibitor displaying 10–20-fold selectivity over Src, c-Raf, CDK2–cyclin A and p38α MAPK, with little inhibition of 20 other protein kinases tested. The compound was also reported to inhibit the LPS-induced production of TNFα (tumour necrosis factor α) in mice, to show efficacy in a model of collagen-induced rheumatoid arthritis and to promote cell survival after cerebral ischaemia [87]. However, when profiled against our panel, AS 601245 was not selective for JNK and inhibited many protein kinases, including p38δ MAPK, ERK8, SGK1, GSK3β, CK2, DYRK1a and PIM isoforms. More detailed kinetic analysis revealed that AS 601245 was an exceptionally potent inhibitor of PIM1, PIM3 and GSK3, with IC50 values in the nanomolar range that were 50–100-fold lower than the IC50 values for JNK1 and JNK2 (Tables 5 and 7).
We recommend that the use of SP 600125 and AS 601245 as JNK inhibitors in cell-based assays be discontinued. The development of a potent and specific inhibitor that can suppress the activities of JNK isoforms in cells would be very useful.
MNK inhibitor CGP 57380
CGP 57380 has been described as an MNK inhibitor [88] and used in cell-based assays for this purpose in several studies. We found that this compound was a relatively weak inhibitor of MNKs, with IC50 values in the low-micromolar range. Against our extended panel, several protein kinases were inhibited with similar potency, including MKK1, CK1 and BRSK2 (Tables 5 and 7). These studies indicate that CGP 57380 is not a specific inhibitor of MNK isoforms and results obtained from its use in cell-based assays are difficult to interpret.
Specificities of some bis(indolyl)maleimides (LY 333531, UCN01, Ro 318220, Go 6976 and KT 5720)
We have previously examined the specificities of a number of bisindolylmaleimides against a smaller panel of protein kinases and found them to inhibit many protein kinases of the AGC subfamily, such as S6K1, RSK2, MSK1 and PKCα [1]. However, at least two of these compounds, UCN01 and LY 333531, have entered clinical trials for the treatment of cancer and diabetic retinopathy respectively, and indeed clinical trials of LY 333531 were only discontinued during Phase III. We therefore studied a few of these compounds against our extended panel (Supplementary Table S2). These studies revealed that LY 333531 was an extremely potent inhibitor of PIM1/PIM3 and RSK1/RSK2, as well as PKCα, and that several other protein kinases were also strongly inhibited, such as PDK1 (see Supplementary Table S2 at http://www.BiochemJ.org/bj/408/bj408ppppadd.htm; for convenience the kinase abbreviations in the footnote are repeated as Supplementary Table S1). UCN01 was an extremely powerful inhibitor of RSK1/RSK2, PRK2, CaMKKβ, PHK, AMPK, MARK3, CHK1, PIM3, MST2 and PDK1, as well as PKCα, while both Ro 318220 and Go 6976 were potent inhibitors of RSK1, RSK2, PRK2, PKCα, PKD1, MSK1, GSK3β, CDK2-cyclin A and PIM3, as well as PKCα. Go 6976 potently inhibited many protein kinases, such as RSK1, CaMKKβ, PHK, CHK1, Aurora B, MST2, and PAKs 4, 5 and 6 (Supplementary Table S2).
KT 5720, which was originally described as an inhibitor of PKA, also inhibited many protein kinases. MKK1, PDK1, PHK, Aurora B and PIM3 were among the protein kinases inhibited most strongly by this compound (Table S2).
In summary, none of the bis(indoyl)maleimides that we have tested are sufficiently specific to be useful as protein kinase inhibitors in cell-based assays.
Protein kinase inhibitor Rottlerin
Rottlerin is a compound extracted from the Monkey-face tree (Mallotus philippinensis Muell.), which grows in the tropical regions of India and has been used for a variety of medicinal purposes over the generations. Although this compound was originally reported to inhibit PKC isoforms, especially PKCδ [89], and has been used as such in many studies, we failed to observe any inhibition of PKCα or PKCδ in a previous study and instead found that MAPKAP-K2 and PRAK were inhibited by this compound [1]. When rottlerin was examined against our extended panel, many more protein kinases were found to inhibited (Supplementary Table S2), those suppressed most strongly being CHK2, PLK1, PIM3 and SRPK1. These observations indicate that rottlerin is too weak and non-specific an inhibitor to be useful in cell-based studies.
Inhibitors of ROCK (H7, H8, HA1077, H89, H1152, Y27632)
Isoquinaline sulphonamide derivatives, such as H7 and H8, developed by Hiroyoshi Hidaka and his colleagues, were among the first inhibitors of protein kinases to be described [90], and the specificities of six of these compounds is shown in Supplementary Table S2. Of these, H89 is marketed as a relatively selective inhibitor of PKA, whereas HA 1077 has been reported to inhibit the Rho-dependent protein kinases PRK1 [91] and ROCK [92], and Y27632 to inhibit ROCK1 and ROCK2 [93]. HA 1077 (also called Fasudil) has been approved in Japan for the treatment of cerebral vasospasm [94], whereas Y27632 has been reported to normalize blood pressure in rodent models of hypertension [93], perhaps by preventing ROCK from inhibiting the major myosin phosphatase in smooth muscle [95]. Y27632 also inhibits RhoA-mediated cell transformation [96], tumour-cell invasion [97] and neutrophil chemotaxis [92], suggesting that inhibitors of ROCK may have therapeutic value as anticancer and anti-inflammatory agents.
We have previously examined the specificities of H89, HA1077 and Y27632 against a panel of 24 protein kinases [1] and here extend the analysis to 70 kinases. H7, H8, H89, HA1077 and H1152 inhibited not only ROCK2 and PRK2, but also other members of the AGC subfamily of protein kinases, such as RSK1, RSK2, PKA and MSK1 with similar or slightly lower potency than ROCK2 and PRK2. The compounds H7, H89 and HA1077 also inhibited AMPK and PKD1, whereas H89 also inhibited PKB isoforms and S6K1, and H-1152 inhibited PHK, Aurora B and Aurora C (Supplementary Table S2). H89 has also been reported to inhibit voltage-dependent potassium ion currents directly by blocking the pore cavity, an effect that was unrelated to the inhibition of PKA [98].
In summary, results obtained by the use of isoquinaline sulphonamides should be interpreted with caution.
Concluding remarks
In the present study we have examined the specificities of many protein kinase inhibitors against a panel of 70–80 protein kinases. The results obtained have re-emphasized the need for great caution in using small-molecule inhibitors of protein kinases to assess the physiological roles of these enzymes. Despite being used widely, many of the compounds analysed in the present study were found to be too non-specific for useful conclusions to be made, other than to exclude the involvement of particular protein kinases in cellular processes. However, extrapolating data obtained from assays performed in vitro to make recommendations about the usefulness of these compounds as inhibitors of particular protein kinases in cells is not straightforward and depends on many factors, such as the stability and cell permeability of the compound, whether it accumulates in the plasma membrane or an intracellular organelle where a particular target is located, the concentration of the protein kinase in vivo and whether the compound is ATP-competitive. The great majority of protein kinase inhibitors that have been developed bind at or near the ATP binding site (PD 184352, PD 0325901, Akt-I-1,2 and rapamycin are exceptions) and, if they were purely ATP-competitive, might be expected to be far less potent in cells where ATP concentrations are in the millimolar range, 100-fold higher than those for assays in vitro. However, this is not always the case, because the specificities of compounds frequently stem from their ability to bind not just in the ATP-binding pocket, but also in neighbouring hydrophobic pockets. Such interactions can induce fast or slow conformational changes and can lock the protein kinase into an inactive state [29,99]. Moreover, some compounds may bind more strongly to the inactive than the active conformation and, like the MKK1 inhibitor PD 98059 [45], prevent the protein kinase from being activated. In these situations the concentration of a compound needed to suppress activity in cells may be similar to, or even lower than, those that inhibit the protein kinase in vitro. Furthermore, a few compounds (such as Akt-I-1,2) only inhibit the full-length protein kinase and not the catalytic domain (the reverse is also theoretically possible). Although catalytic domains are frequently employed for screening purposes if the full-length protein is difficult to express, it should be borne in mind that the use of truncated forms of proteins for screening or to assess specificity may sometimes give misleading results. A further caveat in extrapolating the data obtained in vitro to the cellular context is that only 70–80 protein kinases were used in the present study. Although this is a substantial number, it represents less than 20% of the protein kinases encoded by the human genome. Therefore some compounds may be less specific than the results presented in Tables 1–7 and Supplementary Table S2 would indicate. For this reason, the effects of two structurally unrelated inhibitors of the same protein kinase should be used on cells wherever possible. Off-target effects can also be controlled for by the use of cells that do not express a particular protein kinase or that express a drug-resistant mutant of the protein kinase [27].
Despite the reservations outlined above, a number of compounds were identified that displayed impressive selectivity for a particular protein kinase.
On the basis of the findings reported here and the effects of these compounds on cells that we and our colleagues have studied previously, we recommend that the following compounds be used in cells to assess the roles of particular protein kinases:
For p38α/p38β MAPKs use SB 203580 (1–5 μM) and BIRB 0796 (0.1 μM) in parallel; for substrates of p38γ MAPK, identify proteins whose phosphorylation is prevented by BIRB 0796 (1.0 μM), but not by SB 203580 (5 μM)
For PI3K use PI-103 (0.5 μM) and wortmannin (0.1 μM) in parallel
For PKB/AKT use Akt-I-1, 2 (1 μM)
For MKK1 use PD 184352 (2 μM) or PD 0325901 (0.1 μM)
For TORC1 use rapamycin (100 nM)
For GSK3 use CT 99021 (2 μM)
For RSK use BI-D1870 (10 μM) plus either SL0101 [54] or FMK [53,55]
For CK1 use D4476 (50–100 μM)
VX 680 (1 μM) may be used to inhibit Aurora kinases in cells [64], whereas roscovitine and purvalanol should continue to be useful as pan-CDK inhibitors
Harmine is a very potent and selective inhibitor of DYRK1A, but whether it can be used to assess the role of this protein kinase in cells has yet to be assessed.
It should be noted that some of the recommended inhibitors are not yet available commercially and must be synthesized.
It has been estimated that about 30% of the research and development programmes of the pharmaceutical industry are currently focused on protein kinases (even higher in the field of cancer), and a huge number of compounds with impressive selectivity for particular protein kinases have been developed. The exploitation of these compounds for basic fundamental research by academic scientists is undoubtedly going to be of immense value in the future in facilitating our understanding of the roles of protein kinases in signal-transduction pathways.
Future updates about the specificities of protein kinase inhibitors will appear on the MRC Protein Phosphorylation Unit website (http//www.dundee.ac.uk/lifesciences/mrcppu).
Online data
Acknowledgments
This study was supported by the UK Medical Research Council, The Royal Society, AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck and Co., Merck KGaA and Pfizer. We thank Mark Peggie for site-directed mutagenesis of JNK1, Dr Richard Marais for providing BAY 439006, Dr Morton Frodin for SL0101, Dr Jack Taunton for FMK and Dr Alex Kozikowski for LY 333351.
References
- 1.Davies S. P., Reddy H., Caivano M., Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bain J., McLauchlan H., Elliott M., Cohen P. The specificities of protein kinase inhibitors: an update. Biochem. J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Regan J., Breitfelder S., Cirillo P., Gilmore T., Graham A. G., Hickey E., Klaus B., Madwed J., Moriak M., Moss N., et al. Pyrazole urea-based inhibitors of p38 MAP kinase: from lead compound to clinical candidate. J. Med. Chem. 2002;45:2994–3008. doi: 10.1021/jm020057r. [DOI] [PubMed] [Google Scholar]
- 4.Shpiro N. A., Marquez R. PD-184352. Synth. Commun. 2005;35:2265–2270. [Google Scholar]
- 5.Barrett S. D., Biwersi C., Kaufman M., Tecle H., Warmus J. S. Preparation of oxygenated esters of 4-iodophenylaminobenzhydroxyamic acids as MEK inhibitors. World Pat. WO/2002/006213. 2002
- 6.Nuss J. M., Harrison S. D., Ring D. B., Boyce R. S., Brown S. P., Goff D., Johnson K., Pfister K. B., Ramurthy S., Renhowe P. A., et al. Preparation of aminopyrimidines and pyridines as glycogen synthase kinase 3 inhibitors. World Pat. WO/1999/065897. 1999
- 7.Hoffmann M., Grauert M., Breitfelder S., Eickmeier C., Pohl G., Lehmann-Lintz T., Redemann N., Schnapp G., Steegmaier M., Bauer E., Quant J. J. Preparation of dihydropteridinones as cell proliferation inhibitors. World Pat. WO/2003/020722. 2003
- 8.Bhat R., Xue Y., Berg S., Hellberg S., Ormo M., Nilsson Y., Radesater A. C., Jerning E., Markgren P. O., Borgegard T., et al. Structural insights and biological effects of glycogen synthase kinase 3-specific inhibitor AR-A014418. J. Biol. Chem. 2003;278:45937–45945. doi: 10.1074/jbc.M306268200. [DOI] [PubMed] [Google Scholar]
- 9.Hayakawa M., Kaizawa H., Moritomo H., Kawaguchi K.-I., Koizumi T., Yamano M., Matsuda K., Okada M., Ohta M. Preparation of condensed heteroaryl derivatives as phosphatidylinositol 3-kinase inhibitors and abticancer agents. World Pat. WO/2001/083456. 2001
- 10.Beck J. R. Direct synthesis of benzo[b]thiophene-2-carboxylate esters involving nitro displacement. J. Org. Chem. 1972;37:3224–3226. [Google Scholar]
- 11.Arnautu A., Collot V., Ros J. C., Alayrac C., Witulski B., Rault S. Sonogashira cross coupling reaction of 3-iodoindazoles with various terminal alkynes: a mild and flexible strategy to design 2-azatryptamines. Tetrahedron Lett. 2002;43:2695–2697. [Google Scholar]
- 12.Voisin A. S., Bouillon A., Lancelot J. C., Rault S. Efficient synthesis of halohydroxypyridines by hydroxydeboronation. Tetrahedron. 2005;61:1417–1421. [Google Scholar]
- 13.Li Q., Woods K. W., Zhiu G. D., Fischer J. P., Gong J., Li T., Gandhi V., Thomas S. A., Packard G., Song X., et al. Preparation of pyrimidine derivatives as protein kinases inhibitors. World Pat. WO/2003/051366. 2003
- 14.Burgess J. L., Callahan J. F. Preparation of triarylimidazoles as activin-like kinase (ALK)-5 receptor modulators. World Pat. WO/2000/061576. 2000
- 15.Liverton N. J., Butcher J. W., Claiborne C. F., Claremon D. A., Libby B. E., Nguyen K. T., Pitzenberger S. M., Selnick H. G., Smith G. R., Tebben A., et al. Design and synthesis of potent, selective, and orally bioavailable tetrasubstituted imidazole inhibitors of p38 mitogen-activated protein kinase. J. Med. Chem. 1999;42:2180–2190. doi: 10.1021/jm9805236. [DOI] [PubMed] [Google Scholar]
- 16.Callahan J. F., Burgess J. L., Fornwald J. A., Gaster L. M., Harling J. D., Harrington F. P., Heer J., Kwon C., Lehr R., Mathur A., et al. Identification of novel inhibitors of the transforming growth factor β1 (TGF-β1) type 1 receptor (ALK5) J. Med. Chem. 2002;45:999–1001. doi: 10.1021/jm010493y. [DOI] [PubMed] [Google Scholar]
- 17.Charrier J.-D., Mazzei F., Kay D., Miller A. Processes for preparing 6-pyrazolylpyrimidine as inhibitors of protein kinases, in particular Aurora kinases, by nucleophilic substitution. World Pat. WO/2004/000833. 2004
- 18.Treiber H.-J., Behl B., Hofmann H. P. Preparation of 4-oxoimidazolo[1.2- a]quinoxalines as antagonists of excitatory amino acids. German Pat. DE 4329970. 1994
- 19.Tominaga Y., Honkawa Y., Hara M., Hosomi A. Synthesis of pyrazolo[3,4-D]pyrimidine derivatives using ketene dithioacetate. J. Heterocyclic Chem. 1990;27:775–783. [Google Scholar]
- 20.Bryant J., Kochanny M. J., Yuan S., Khim S. K., Buckman B., Arnaiz D., Boemer U., Briem H., Esperling P., Huwe P., et al. Chk-, Pdk- and Akt-inhibitory pyrimidines, their production and use as pharmaceutical agents. World Pat. WO/2004/048343. 2004
- 21.Sun L., Tran N., Liang C., Tang F., Rice A., Schreck R., Waltz K., Shawver L. K., McMahon G., Tang C. Design, synthesis, and evaluations of substituted 3-[(3- or 4-carboxyethylpyrrol-2-yl)methylidenyl]indolin-2-ones as inhibitors of VEGF, FGF, and PDGF receptor tyrosine kinases. J. Med. Chem. 1999;42:5120–5130. doi: 10.1021/jm9904295. [DOI] [PubMed] [Google Scholar]
- 22.Alessi D. R., Cohen P., Ashworth A., Cowley S., Leevers S. J., Marshall C. J. Assay and expression of mitogen-activated protein kinase, MAP kinase kinase, and Raf. Methods Enzymol. 1995;255:279–290. doi: 10.1016/s0076-6879(95)55031-3. [DOI] [PubMed] [Google Scholar]
- 23.Cuenda A., Rouse J., Doza Y. N., Meier R., Cohen P., Gallagher T. F., Young P. R., Lee J. C. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 24.Godl K., Wissing J., Kurtenbach A., Habenberger P., Blencke S., Gutbrod H., Salassidis K., Stein-Gerlach M., Missio A., Cotten M., Daub H. An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15434–15439. doi: 10.1073/pnas.2535024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall-Jackson C. A., Goedert M., Hedge P., Cohen P. Effect of SB 203580 on the activity of c-Raf in vitro and in vivo. Oncogene. 1999;18:2047–2054. doi: 10.1038/sj.onc.1202603. [DOI] [PubMed] [Google Scholar]
- 26.Fryer L. G., Parbu-Patel A., Carling D. Protein kinase inhibitors block the stimulation of the AMP-activated protein kinase by 5-amino-4-imidazolecarboxamide riboside. FEBS Lett. 2002;531:189–192. doi: 10.1016/s0014-5793(02)03501-9. [DOI] [PubMed] [Google Scholar]
- 27.Eyers P. A., van den IJssel P., Quinlan R. A., Goedert M., Cohen P. Use of a drug-resistant mutant of stress-activated protein kinase 2a/p38 to validate the in vivo specificity of SB 203580. FEBS Lett. 1999;451:191–196. doi: 10.1016/s0014-5793(99)00552-9. [DOI] [PubMed] [Google Scholar]
- 28.Cheung P. C., Campbell D. G., Nebreda A. R., Cohen P. Feedback control of the protein kinase TAK1 by SAPK2a/p38α. EMBO J. 2003;22:5793–5805. doi: 10.1093/emboj/cdg552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pargellis C., Tong L., Churchill L., Cirillo P. F., Gilmore T., Graham A. G., Grob P. M., Hickey E. R., Moss N., Pav S., Regan J. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat. Struct. Biol. 2002;9:268–272. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- 30.Kuma Y., Sabio G., Bain J., Shpiro N., Marquez R., Cuenda A. BIRB796 inhibits all p38 MAPK isoforms in vitro and in vivo. J. Biol. Chem. 2005;280:19472–19479. doi: 10.1074/jbc.M414221200. [DOI] [PubMed] [Google Scholar]
- 31.Jaeschke A., Karasarides M., Ventura J. J., Ehrhardt A., Zhang C., Flavell R. A., Shokat K. M., Davis R. J. JNK2 is a positive regulator of the cJun transcription factor. Mol. Cell. 2006;23:899–911. doi: 10.1016/j.molcel.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 32.Ventura J. J., Hubner A., Zhang C., Flavell R. A., Shokat K. M., Davis R. J. Chemical genetic analysis of the time course of signal transduction by JNK. Mol. Cell. 2006;21:701–710. doi: 10.1016/j.molcel.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 33.Blake R. A., Broome M. A., Liu X., Wu J., Gishizky M., Sun L., Courtneidge S. A. SU6656, a selective src family kinase inhibitor, used to probe growth factor signaling. Mol. Cell Biol. 2000;20:9018–9027. doi: 10.1128/mcb.20.23.9018-9027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanke J. H., Gardner J. P., Dow R. L., Changelian P. S., Brissette W. H., Weringer E. J., Pollok B. A., Connelly P. A. Discovery of a novel, potent, and Src family-selective tyrosine kinase inhibitor. Study of Lck- and FynT-dependent T cell activation. J. Biol. Chem. 1996;271:695–701. doi: 10.1074/jbc.271.2.695. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y., Bishop A., Witucki L., Kraybill B., Shimizu E., Tsien J., Ubersax J., Blethrow J., Morgan D. O., Shokat K. M. Structural basis for selective inhibition of Src family kinases by PP1. Chem. Biol. 1999;6:671–678. doi: 10.1016/s1074-5521(99)80118-5. [DOI] [PubMed] [Google Scholar]
- 36.Windheim M., Lang C., Peggie M., Cummings L. A., Cohen P. Molecular mechanisms involved in the regulation of cytokine production by muramyl dipeptide. Biochem. J. 2007;404:179–190. doi: 10.1042/BJ20061704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eyers P. A., Craxton M., Morrice N., Cohen P., Goedert M. Conversion of SB 203580-insensitive MAP kinase family members to drug-sensitive forms by a single amino-acid substitution. Chem. Biol. 1998;5:321–328. doi: 10.1016/s1074-5521(98)90170-3. [DOI] [PubMed] [Google Scholar]
- 38.Gum R. J., McLaughlin M. M., Kumar S., Wang Z., Bower M. J., Lee J. C., Adams J. L., Livi G. P., Goldsmith E. J., Young P. R. Acquisition of sensitivity of stress-activated protein kinases to the p38 inhibitor, SB 203580, by alteration of one or more amino acids within the ATP binding pocket. J. Biol. Chem. 1998;273:15605–15610. doi: 10.1074/jbc.273.25.15605. [DOI] [PubMed] [Google Scholar]
- 39.Bishop A. C., Ubersax J. A., Petsch D. T., Matheos D. P., Gray N. S., Blethrow J., Shimizu E., Tsien J. Z., Schultz P. G., Rose M. D., et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 40.Wiggin G. R., Soloaga A., Foster J. M., Murray-Tait V., Cohen P., Arthur J. S. MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol. Cell Biol. 2002;22:2871–2881. doi: 10.1128/MCB.22.8.2871-2881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaw M., Cohen P. Role of protein kinase B and the MAP kinase cascade in mediating the EGF-dependent inhibition of glycogen synthase kinase 3 in Swiss 3T3 cells. FEBS Lett. 1999;461:120–124. doi: 10.1016/s0014-5793(99)01434-9. [DOI] [PubMed] [Google Scholar]
- 42.Hall-Jackson C. A., Eyers P. A., Cohen P., Goedert M., Boyle F. T., Hewitt N., Plant H., Hedge P. Paradoxical activation of Raf by a novel Raf inhibitor. Chem. Biol. 1999;6:559–568. doi: 10.1016/s1074-5521(99)80088-x. [DOI] [PubMed] [Google Scholar]
- 43.Lyons J. F., Wilhelm S., Hibner B., Bollag G. Discovery of a novel Raf kinase inhibitor. Endocr. Relat. Cancer. 2001;8:219–225. doi: 10.1677/erc.0.0080219. [DOI] [PubMed] [Google Scholar]
- 44.Wilhelm S. M., Carter C., Tang L., Wilkie D., McNabola A., Rong H., Chen C., Zhang X., Vincent P., McHugh M., et al. BAY 43–9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64:7099–7109. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- 45.Alessi D. R., Cuenda A., Cohen P., Dudley D. T., Saltiel A. R. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J. Biol. Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 46.Favata M. F., Horiuchi K. Y., Manos E. J., Daulerio A. J., Stradley D. A., Feeser W. S., Van Dyk D. E., Pitts W. J., Earl R. A., Hobbs F., et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 47.Sebolt-Leopold J. S., Dudley D. T., Herrera R., Van Becelaere K., Wiland A., Gowan R. C., Tecle H., Barrett S. D., Bridges A., Przybranowski S., et al. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat. Med. 1999;5:810–816. doi: 10.1038/10533. [DOI] [PubMed] [Google Scholar]
- 48.Reference deleted
- 49.Kamakura S., Moriguchi T., Nishida E. Activation of the protein kinase ERK5/BMK1 by receptor tyrosine kinases. Identification and characterization of a signaling pathway to the nucleus. J. Biol. Chem. 1999;274:26563–26571. doi: 10.1074/jbc.274.37.26563. [DOI] [PubMed] [Google Scholar]
- 50.Mody N., Leitch J., Armstrong C., Dixon J., Cohen P. Effects of MAP kinase cascade inhibitors on the MKK5/ERK5 pathway. FEBS Lett. 2001;502:21–24. doi: 10.1016/s0014-5793(01)02651-5. [DOI] [PubMed] [Google Scholar]
- 51.Sapkota G. P., Cummings L., Newell F. S., Armstrong C., Bain J., Frodin M., Grauert M., Hoffmann M., Schnapp G., Steegmaier M., et al. BI-D1870 is a specific inhibitor of the p90 RSK (ribosomal S6 kinase) isoforms in vitro and in vivo. Biochem. J. 2007;401:29–38. doi: 10.1042/BJ20061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith J. A., Poteet-Smith C. E., Xu Y., Errington T. M., Hecht S. M., Lannigan D. A. Identification of the first specific inhibitor of p90 ribosomal S6 kinase (RSK) reveals an unexpected role for RSK in cancer cell proliferation. Cancer Res. 2005;65:1027–1034. [PubMed] [Google Scholar]
- 53.Cohen M. S., Zhang C., Shokat K. M., Taunton J. Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science. 2005;308:1318–1321. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zaru R., Ronkina N., Gaestel M., Arthur J. S. C., Watts C. The MAP kinase-activated Rsk controls an acute Toll-like receptor signalling response in dendritic cells and is activated via two distinct pathways. Nat. Immunol. 2007;8:1227–1235. doi: 10.1038/ni1517. [DOI] [PubMed] [Google Scholar]
- 55.Cohen M. S., Hadjivassiliou H., Taunton J. A clickable inhibitor reveals context-dependent autoactivation of p90 RSK. Nat. Chem. Biol. 2007;3:156–160. doi: 10.1038/nchembio859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen P., Goedert M. GSK3 inhibitors: development and therapeutic potential. Nat. Rev. Drug Discov. 2004;3:479–487. doi: 10.1038/nrd1415. [DOI] [PubMed] [Google Scholar]
- 57.Murray J. T., Campbell D. G., Morrice N., Auld G. C., Shpiro N., Marquez R., Peggie M., Bain J., Bloomberg G. B., Grahammer F., et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem. J. 2004;384:477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakanishi S., Kakita S., Takahashi I., Kawahara K., Tsukuda E., Sano T., Yamada K., Yoshida M., Kase H., Matsuda Y., et al. Wortmannin, a microbial product inhibitor of myosin light chain kinase. J. Biol. Chem. 1992;267:2157–2163. [PubMed] [Google Scholar]
- 59.Liu Y., Shreder K. R., Gai W., Corral S., Ferris D. K., Rosenblum J. S. Wortmannin, a widely used phosphoinositide 3-kinase inhibitor, also potently inhibits mammalian polo-like kinase. Chem. Biol. 2005;12:99–107. doi: 10.1016/j.chembiol.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 60.Gharbi S. I., Zvelebil M. J., Shuttleworth S. J., Hancox T., Saghir N., Timms J. F., Waterfield M. D. Exploring the specificity of the PI3K family inhibitor LY294002. Biochem. J. 2007;404:15–21. doi: 10.1042/BJ20061489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Workman P., Raynaud F., Clarke P., te Poele R., Eccles S., Kelland L., Di Stefano F., Ahmadi K., Parker P., Waterfield M. D. Pharmacological properties and in vitro and in vivo antitumour activity of the potent and selective PI3 kinase inhibitor PI103. Eur. J. Cancer. 2004;40:414A. [Google Scholar]
- 62.Fan Q. W., Knight Z. A., Goldenberg D. D., Yu W., Mostov K. E., Stokoe D., Shokat K. M., Weiss W. A. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell. 2006;9:341–349. doi: 10.1016/j.ccr.2006.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heitman J., Movva N. R., Hall M. N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 64.Tyler R. K., Shpiro N., Marquez R., Eyers P. A. VX-680 inhibits Aurora A and Aurora B kinase activity in human cells. Cell Cycle. 2007;6:in the press. doi: 10.4161/cc.6.22.4940. [DOI] [PubMed] [Google Scholar]
- 65.Bayascas J. R., Leslie N. R., Parsons R., Fleming S., Alessi D. R. Hypomorphic mutation of PDK1 suppresses tumorigenesis in PTEN+/− mice. Curr. Biol. 2005;15:1839–1846. doi: 10.1016/j.cub.2005.08.066. [DOI] [PubMed] [Google Scholar]
- 66.Feldman R. I., Wu J. M., Polokoff M. A., Kochanny M. J., Dinter H., Zhu D., Biroc S. L., Alicke B., Bryant J., Yuan S., et al. Novel small molecule inhibitors of 3-phosphoinositide-dependent kinase-1. J. Biol. Chem. 2005;280:19867–19874. doi: 10.1074/jbc.M501367200. [DOI] [PubMed] [Google Scholar]
- 67.Korherr C., Gille H., Schafer R., Koenig-Hoffmann K., Dixelius J., Egland K. A., Pastan I., Brinkmann U. Identification of proangiogenic genes and pathways by high-throughput functional genomics: TBK1 and the IRF3 pathway. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4240–4245. doi: 10.1073/pnas.0511319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chien Y., Kim S., Bumeister R., Loo Y. M., Kwon S. W., Johnson C. L., Balakireva M. G., Romeo Y., Kopelovich L., Gale M., Jr., et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 69.Luo Y., Shoemaker A. R., Liu X., Woods K. W., Thomas S. A., de Jong R., Han E. K., Li T., Stoll V. S., Powlas J. A., et al. Potent and selective inhibitors of Akt kinases slow the progress of tumors in vivo. Mol. Cancer Ther. 2005;4:977–986. doi: 10.1158/1535-7163.MCT-05-0005. [DOI] [PubMed] [Google Scholar]
- 70.Barnett S. F., Defeo-Jones D., Fu S., Hancock P. J., Haskell K. M., Jones R. E., Kahana J. A., Kral A. M., Leander K., Lee L. L., et al. Identification and characterization of pleckstrin-homology-domain-dependent and isoenzyme-specific Akt inhibitors. Biochem. J. 2005;385:399–408. doi: 10.1042/BJ20041140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Logie L., Ruiz-Alcaraz A. J., Keane M., Woods Y. L., Bain J., Marquez R., Alessi D. R., Sutherland C. Characterisation of a protein kinase B inhibitor in vitro and in insulin treated liver cells. Diabetes. 2007;56:2218–2227. doi: 10.2337/db07-0343. [DOI] [PubMed] [Google Scholar]
- 72.Rena G., Bain J., Elliott M., Cohen P. D4476, a cell-permeant inhibitor of CK1, suppresses the site-specific phosphorylation and nuclear exclusion of FOXO1a. EMBO Rep. 2004;5:60–65. doi: 10.1038/sj.embor.7400048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song Y., Kesuma D., Wang J., Deng Y., Duan J., Wang J. H., Qi R. Z. Specific inhibition of cyclin-dependent kinases and cell proliferation by harmine. Biochem. Biophys. Res. Commun. 2004;317:128–132. doi: 10.1016/j.bbrc.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 74.Altafaj X., Dierssen M., Baamonde C., Marti E., Visa J., Guimera J., Oset M., Gonzalez J. R., Florez J., Fillat C., Estivill X. Neurodevelopmental delay, motor abnormalities and cognitive deficits in transgenic mice overexpressing Dyrk1A (minibrain), a murine model of Down's syndrome. Hum. Mol. Genet. 2001;10:1915–1923. doi: 10.1093/hmg/10.18.1915. [DOI] [PubMed] [Google Scholar]
- 75.Arron J. R., Winslow M. M., Polleri A., Chang C. P., Wu H., Gao X., Neilson J. R., Chen L., Heit J. J., Kim S. K., et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- 76.Waki H., Park K. W., Mitro N., Pei L., Damoiseaux R., Wilpitz D. C., Reue K., Saez E., Tontonoz P. The small molecule harmine is an antidiabetic cell-type-specific regulator of PPARγ expression. Cell Metab. 2007;5:357–370. doi: 10.1016/j.cmet.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 77.Meijer L., Borgne A., Mulner O., Chong J. P., Blow J. J., Inagaki N., Inagaki M., Delcros J. G., Moulinoux J. P. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur. J. Biochem. 1997;243:527–536. doi: 10.1111/j.1432-1033.1997.t01-2-00527.x. [DOI] [PubMed] [Google Scholar]
- 78.Gray N. S., Wodicka L., Thunnissen A. M., Norman T. C., Kwon S., Espinoza F. H., Morgan D. O., Barnes G., LeClerc S., Meijer L., et al. Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science. 1998;281:533–538. doi: 10.1126/science.281.5376.533. [DOI] [PubMed] [Google Scholar]
- 79.Bach S., Knockaert M., Reinhardt J., Lozach O., Schmitt S., Baratte B., Koken M., Coburn S. P., Tang L., Jiang T. Roscovitine targets, protein kinases and pyridoxal kinase. J. Biol. Chem. 2005;280:31208–31219. doi: 10.1074/jbc.M500806200. [DOI] [PubMed] [Google Scholar]
- 80.Carter T. A., Wodicka L. M., Shah N. P., Velasco A. M., Fabian M. A., Treiber D. K., Milanov Z. V., Atteridge C. E., Biggs W. H., 3rd, Edeen P. T., et al. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11011–11016. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harrington E. A., Bebbington D., Moore J., Rasmussen R. K., Ajose-Adeogun A. O., Nakayama T., Graham J. A., Demur C., Hercend T., Diu-Hercend A., et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat. Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 82.Godl K., Gruss O. J., Eickhoff J., Wissing J., Blencke S., Weber M., Degen H., Brehmer D., Orfi L., Horvath Z., et al. Proteomic characterization of the angiogenesis inhibitor SU6668 reveals multiple impacts on cellular kinase signaling. Cancer Res. 2005;65:6919–6926. doi: 10.1158/0008-5472.CAN-05-0574. [DOI] [PubMed] [Google Scholar]
- 83.Hawley S. A., Pan D. A., Mustard K. J., Ross L., Bain J., Edelman A. M., Frenguelli B. G., Hardie D. G. Calmodulin-dependent protein kinase kinase-β is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2005;2:9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 84.Tamas P., Hawley S. A., Clarke R. G., Mustard K. J., Green K., Hardie D. G., Cantrell D. A. Regulation of the energy sensor AMP-activated protein kinase by antigen receptor and Ca2+ in T lymphocytes. J. Exp. Med. 2006;203:1665–1670. doi: 10.1084/jem.20052469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cho J., Melnick M., Solidakis G. P., Tsichlis P. N. Tpl2 (tumor progression locus 2) phosphorylation at Thr290 is induced by lipopolysaccharide via an IκB kinase-β-dependent pathway and is required for Tpl2 activation by external signals. J. Biol. Chem. 2005;280:20442–20448. doi: 10.1074/jbc.M413554200. [DOI] [PubMed] [Google Scholar]
- 86.Stafford M. J., Morrice N. A., Peggie M. W., Cohen P. Interleukin-1 stimulated activation of the COT catalytic subunit through the phosphorylation of Thr290 and Ser62. FEBS Lett. 2006;580:4010–4014. doi: 10.1016/j.febslet.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 87.Gaillard P., Jeanclaude-Etter I., Ardissone V., Arkinstall S., Cambet Y., Camps M., Chabert C., Church D., Cirillo R., Gretener D. Design and synthesis of the first generation of novel potent, selective, and in vivo active (benzothiazol-2-yl)acetonitrile inhibitors of the c-Jun N-terminal kinase. J. Med. Chem. 2005;48:4596–4607. doi: 10.1021/jm0310986. [DOI] [PubMed] [Google Scholar]
- 88.Knauf U., Tschopp C., Gram H. Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2. Mol. Cell Biol. 2001;21:5500–5511. doi: 10.1128/MCB.21.16.5500-5511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gschwendt M., Muller H. J., Kielbassa K., Zang R., Kittstein W., Rincke G., Marks F. Rottlerin, a novel protein kinase inhibitor. Biochem. Biophys. Res. Commun. 1994;199:93–98. doi: 10.1006/bbrc.1994.1199. [DOI] [PubMed] [Google Scholar]
- 90.Hidaka H., Inagaki M., Kawamoto S., Sasaki Y. Isoquinolinesulfonamides, novel and potent inhibitors of cyclic nucleotide dependent protein kinase and protein kinase C. Biochemistry. 1984;23:5036–5041. doi: 10.1021/bi00316a032. [DOI] [PubMed] [Google Scholar]
- 91.Amano M., Chihara K., Nakamura N., Kaneko T., Matsuura Y., Kaibuchi K. The COOH terminus of Rho-kinase negatively regulates rho-kinase activity. J. Biol. Chem. 1999;274:32418–32424. doi: 10.1074/jbc.274.45.32418. [DOI] [PubMed] [Google Scholar]
- 92.Niggli V. Rho-kinase in human neutrophils: a role in signalling for myosin light chain phosphorylation and cell migration. FEBS Lett. 1999;445:69–72. doi: 10.1016/s0014-5793(99)00098-8. [DOI] [PubMed] [Google Scholar]
- 93.Uehata M., Ishizaki T., Satoh H., Ono T., Kawahara T., Morishita T., Tamakawa H., Yamagami K., Inui J., Maekawa M., Narumiya S. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389:990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- 94.Tachibana E., Harada T., Shibuya M., Saito K., Takayasu M., Suzuki Y., Yoshida J. Intra-arterial infusion of fasudil hydrochloride for treating vasospasm following subarachnoid haemorrhage. Acta Neurochir. 1999;141:13–19. doi: 10.1007/s007010050260. [DOI] [PubMed] [Google Scholar]
- 95.Kimura K., Ito M., Amano M., Chihara K., Fukata Y., Nakafuku M., Yamamori B., Feng J., Nakano T., Okawa K., et al. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 96.Sahai E., Ishizaki T., Narumiya S., Treisman R. Transformation mediated by RhoA requires activity of ROCK kinases. Curr. Biol. 1999;9:136–145. doi: 10.1016/s0960-9822(99)80067-0. [DOI] [PubMed] [Google Scholar]
- 97.Itoh K., Yoshioka K., Akedo H., Uehata M., Ishizaki T., Narumiya S. An essential part for Rho-associated kinase in the transcellular invasion of tumor cells. Nat. Med. 1999;5:221–225. doi: 10.1038/5587. [DOI] [PubMed] [Google Scholar]
- 98.Son Y. K., Park W. S., Kim S. J., Earm Y. E., Kim N., Youm J. B., Warda M., Kim E., Han J. Direct inhibition of a PKA inhibitor, H-89 on KV channels in rabbit coronary arterial smooth muscle cells. Biochem. Biophys. Res. Commun. 2006;341:931–937. doi: 10.1016/j.bbrc.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 99.Schindler T., Bornmann W., Pellicena P., Miller W. T., Clarkson B., Kuriyan J. Structural mechanism for STI-571 inhibition of Abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.