Abstract
Substantial data indicate that microRNA 21 (miR-21) is significantly elevated in glioblastoma (GBM) and in many other tumors of various origins. This microRNA has been implicated in various aspects of carcinogenesis, including cellular proliferation, apoptosis, and migration. We demonstrate that miR-21 regulates multiple genes associated with glioma cell apoptosis, migration, and invasiveness, including the RECK and TIMP3 genes, which are suppressors of malignancy and inhibitors of matrix metalloproteinases (MMPs). Specific inhibition of miR-21 with antisense oligonucleotides leads to elevated levels of RECK and TIMP3 and therefore reduces MMP activities in vitro and in a human model of gliomas in nude mice. Moreover, downregulation of miR-21 in glioma cells leads to decreases of their migratory and invasion abilities. Our data suggest that miR-21 contributes to glioma malignancy by downregulation of MMP inhibitors, which leads to activation of MMPs, thus promoting invasiveness of cancer cells. Our results also indicate that inhibition of a single oncomir, like miR-21, with specific antisense molecules can provide a novel therapeutic approach for “physiological” modulation of multiple proteins whose expression is deregulated in cancer.
Malignant gliomas are brain tumors of glial origin. They are the most common type of primary brain tumors in adults and persist as serious clinical and scientific problems (reviewed in reference 40). Survival depends heavily on the histological grade of the tumor, but patients afflicted with the most malignant glioma, glioblastoma (GBM), survive on average less than 1 year. Current therapies for GBM, though they are very aggressive and usually include surgery, radiotherapy, and chemotherapy, have not been successful, due to several factors. These include rapidness and invasiveness of tumor growth, the genetic heterogeneity of the tumors, and our poor understanding of the molecular mechanisms governing disease manifestation and progression (40).
MicroRNAs (miRNAs) are small regulatory RNA molecules that in recent years have been identified in the progression of various cancers and proposed as novel targets for anticancer therapies (reviewed in references 9 and 13). By negatively regulating their mRNA targets to either degradation or translational repression, they can act as both tumor suppressors and oncogenes (19, 27, 41, 43). Using high-throughput profiling of miRNA expression, we have previously identified a specific miRNA, miRNA 21 (miR-21), as most strongly elevated in nearly all analyzed human GBM specimens (5). Other groups demonstrated overexpression of this miRNA in a wide range of other cancers, including breast, lung, colon, prostate, pancreas, ovarian, and stomach cancers, as well as in chronic lymphocytic leukemia (33, 54). These combined findings suggest miR-21 as a possible oncogene acting in a variety of cancers. miR-21 has been identified in controlling apoptosis, cell proliferation, and migration of cell lines in breast, colorectal, and other cancers (1, 44, 51, 59).
Our aim was to identify the major miR-21 targets and signaling pathways mediating its function in gliomas. In animal and human cells, miRNAs share only partial complementarity to their targets, and the conditions required for miRNA targeting have not been fully established. Therefore, identification and validation of the key targets that function in a specific cell setting or process is a challenge. Various studies indicate the importance of the 5′ end of the miRNA (the first 2 to 8 nucleotides, called the “seed”) for proper mRNA recognition and targeting function (34, 55). Other determinants of functional targeting include the nucleotide composition around the binding site, the position within the 3′ untranslated region (UTR), and the complementarity at the 3′ end of the miRNA (17). Base pairing at the 5′ seed region of the miRNA appears to be the strongest indicator of targeting. One widely used approach for target identification relies on detection of mRNAs whose expression levels are modulated by exogenously added miRNA mimics or inhibitors (36, 37). We used this approach to discover miR-21 targets and function in glioma cells in vitro and a human glioma model in nude mice in vivo.
MATERIALS AND METHODS
Human tissue samples.
Fresh frozen human nonneoplastic brain tissue and human tumor samples were obtained from the Department of Pathology at Brigham and Women's Hospital. All human materials were used in accordance with the policies of the institutional review board at Brigham and Women's Hospital.
Cell cultures and oligonucleotide transfections.
The human A172, U87, and LN229 glioma cell lines, which express high levels of miR-21 (5), were used for in vitro and in vivo experiments. A172, U87, LN229, cervix carcinoma HeLa, breast carcinoma MCF7, and human osteosarcoma U2OS cells were obtained from the ATCC. The cells were maintained in either Dulbecco's modified Eagle's medium (DMEM) (U87, A172, LN229, HeLa, and U2OS cells) or RPMI medium supplemented with 10% fetal calf serum (MCF7 cells). For miR-21 inhibition, the cells were transfected with 50 nM of 2′-O-methoxyethyl (2′-O-MOE) oligonucleotides by Lipofectamine 2000 (Invitrogen), according to the manufacturer's protocol. 2′-O-MOE oligonucleotides with phosphate backbone were synthesized as previously described (10). The 2′-O-MOE sequences were as follows: for the miR-21 antisense oligonucleotide (ASO), 5′-TCAACATCAGTCTGATAAGCTA-3′; for the mismatch oligonucleotide, 5′-TCTTCATGAGTCAGATTACCTA-3′; and for the control oligonucleotide, 5′-ACATACTCCTTTCTCAGAGTCCA-3′. The sequences of the control and mismatch oligonucleotides have been analyzed using BLAST searches to exclude potential hits in the human transcriptome. For overexpression, the cells were transfected with a synthetic RNA duplex (obtained from IDT) corresponding to mature miR-21. For RECK RNA interference, the following sense sequence was used for targeting: 5′-GACCAGCCCUUGCCUCAAUU-3′.
mRNA arrays and computational data analysis.
A172 glioma cells treated with anti-miR-21, mismatch control antagomiR, or the miR-21 mimetic duplex were compared to mock-treated A172 glioma cells as a reference. Cells were harvested at 6 and 24 h posttreatment, comprising six pairs of samples in all. Agilent microarrays were hybridized as fluor-reversed pairs of arrays.
Microarray analysis was performed as described previously (22, 24). Regulated transcripts were identified in microarray gene expression signatures, using a P value cutoff (P < 0.05). Regulated transcripts were tested for enrichment relative to a background set, using hypergeometric distribution. miR-21 target regulation was measured by enrichment of transcripts containing miR-21 hexamer seed strings (stretches of 6 contiguous bases complementary to miRNA seed region nucleotides 1 to 6, 2 to 7, or 3 to 8) in transcripts having annotated 3′ UTRs. Biological function was detected by enrichment of transcripts from Gene Ontology database biological process functional categories (http://www.geneontology.org/). The set of genes represented by probes on the microarray was used as a background set.
qRT-PCR analysis of mRNA and miRNA expression.
Total RNA from tissues and cells was isolated using TRIzol reagent (Invitrogen) for both mRNA and miRNA analyses. Relative levels of mRNA were examined using Sybr green real-time quantitative reverse transcription-PCR (qRT-PCR) (Applied Biosystems) and normalized to levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. For analysis of miRNA expression, real-time RT-PCR analyses were carried out using TaqMan miRNA assays (Applied Biosystems) according to the manual. Relative expression was calculated using the ΔΔCT method (38) and normalized to the expression of snoRNA48 (Applied Biosystems) or relatively uniformly expressed miR-19b. All qRT-PCRs were performed in duplicate, and the data are presented as means ± standard errors of the means (SEM).
Western blot analysis.
Cells were transfected with either anti-miR-21 or control oligonucleotides in tissue culture plates and grown for 72 h. The cells were lysed by a standard procedure in radioimmunoprecipitation assay buffer containing protease inhibitor cocktail (Roche Diagnostics). Protein concentrations of total cell lysates were measured using a Micro BCA protein assay kit (Pierce Biotechnology), and 50 μg per lane of total cell lysates was resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (Invitrogen), followed by immunoblot detection and visualization with ECL Western blotting detection reagents (Pierce Biotechnology). Immunoblotting was performed with the following primary antibodies: mouse anti-RECK (1:250; BD Biosciences Pharmingen), rabbit anti-TIMP3 (1:1,000; Chemicon International, Inc.), and mouse antiactin (1:5,000; Abcam).
Luciferase miRNA target reporter assay.
Total cDNA from A172 cells was used to amplify the 3′ UTRs of RECK and TIMP3 by PCR using the forward primers ATTAACTAGTACCTCTATTCGCCACACAG and TATGACTAGTAGCCCAGTGATGCTTGTGTTG and the reverse primers CTACATCAGCACTGACATATTCTG and TATGAAGCTTATTCAGGAAAATGGCGGCATGTG, correspondingly. After digestion of the PCR product by SpeI and HindIII, the RECK and TIMP3 3′ UTRs were cloned into the SpeI and HindIII sites of pMir-Report (Ambion), resulting in pMir-Report-3′RECK and pMir-Report-3′TIMP3. Mutations were introduced in the potential miR-21 binding sites by using a QuikChange site-directed mutagenesis kit (Stratagene). HeLa and A172 cells were transfected with the pMir-Report vectors containing the 3′ UTR variants, and at 5 h after transfection, the cells were transfected again with 50 nM of anti-miR-21 or control oligonucleotides. After 24 h, the cells were lysed and luciferase activity was measured. pRenilla was cotransfected and used for normalization.
Scratch migration assay.
Confluent A172 cells were transfected with anti-miR-21 inhibitors or control oligonucleotides. At 5 h after transfection, a cell scratch spatula was used to make a scratch in the cell monolayer. Pictures of the scratches were taken (×4 magnification) by using a digital camera system coupled with a microscope. The cells were incubated for 24 to 72 h, and pictures were taken at different times. The software program MetaVue was used to determine migration distance (in micrometers).
Zymography.
To measure the activity of matrix metalloproteinase (MMP) in glioma cells (A172 and U87), 15 μl of conditioned medium was mixed 1:1 with Zymogram sample buffer (Bio-Rad Laboratories) and resolved on 10% Novex Zymogram gels (Invitrogen). Following electrophoresis, gels were incubated at room temperature in Zymogram renaturing buffer (Bio-Rad Laboratories) for 30 min. Next, the gels were equilibrated in Zymogram development buffer (Bio-Rad Laboratories) at room temperature for 30 min and then incubated at 37°C overnight. The gels were washed and stained in Imperial protein stain (Pierce) according to the manufacturer's protocol. Quantification analysis was performed using the software program ImageJ (NIH).
In vitro invasion assay.
A172 and U87 cells were transfected with either anti-miR-21 or control oligonucleotides, cultured for 48 h, and transferred on the top of Matrigel-coated invasion chambers (24-well insert, 8-μm pore size; BD Biosciences) in a serum-free DMEM. As a chemoattractant, DMEM containing 10% fetal calf serum was added to the lower chamber. After incubation for 20 h, noninvaded cells were removed from the inner part of the insert by using a cotton swab. Fixation and staining of invaded cells were performed using a Diff-Quick differential staining set (Dade Behring, Inc.).
In vivo imaging of MMP activity.
U87 cells were transfected with 50 nM anti-miR-21 inhibitors or control oligonucleotides. At 24 h after transfection, 150 μl phosphate-buffered saline containing 1 × 106 cells was mixed with 50 μl Matrigel (Becton Dickinson) and directly injected subcutaneously in the flanks of nude mice. Each mouse was injected with U87 cells transfected with antisense anti-miR-21 (left flank) and control oligonucleotide U87 cells (right flank). At 3 days after injection of the cells, 150 μl of MMPsense 680 (Visen) was injected intravenously. After 24 h, the mice were subjected to fluorescence imaging using a Kodak fluorescence imaging system (32). The relative fluorescence intensities were determined using ImageJ software (NIH). In parallel, caliper measurements were performed to track tumor growth.
Microarray data accession number.
The microarray data have been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/projects/geo/) and assigned accession number GSE11778.
RESULTS
miR-21 expression correlates with glioma grade.
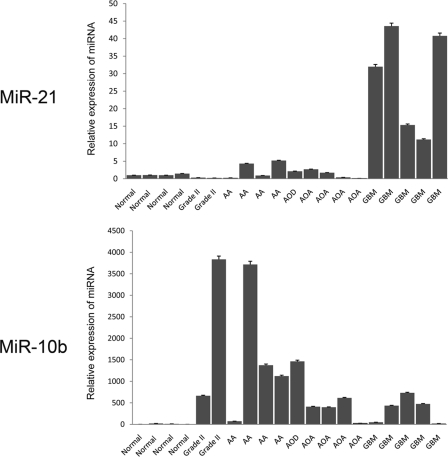
We have previously demonstrated that miR-21 expression is upregulated in GBMs (5). To further assess the relevance of miR-21 in glioma tumorigenicity, we determined miR-21 levels in tumors of different grades (grade II [low-grade gliomas], grade III [anaplastic astrocytomas and oligodendrogliomas], and grade IV [GBMs]) compared to normal brain tissue by qRT-PCR. miR-21 levels remain low in grade II gliomas and in most grade III tumors and are significantly higher in GBMs (Fig. 1). This expression pattern is different from those of the other cancer-associated miRNAs, for example, miR-10b, which is often elevated in grade III and even grade II gliomas (Fig. 1) and dysregulated in some other cancers (41). The most important characteristics used for glioma grading and GBM diagnostics are microvascular proliferation, which is associated with reorganization of the extracellular matrix (ECM), and high proliferative capacity in the tumor cells (40). The high level of miR-21 expression in GBMs compared to that in anaplastic astrocytomas suggests the involvement of miR-21 in one of these key processes.
FIG. 1.
miR-21 expression in glioma progression. qRT-PCRs for miR-21 and miR-10b have been performed with primers specific for mature miRNAs and normalized to levels for miR-19b uniformly expressed among control brain tissues and tumors. Normal brain tissues (Normal) and gliomas (grade II, grade III [AA, anaplastic astrocytoma; AOD, anaplastic oligodendroglioma; AOA, anaplastic oligoastrocytoma], grade IV [GBM]) were examined. The reactions were performed in duplicates, and the data are represented as means ± SEM.
mRNA profiling of miR-21-modulated gliomas: apoptosis and ECM remodeling.
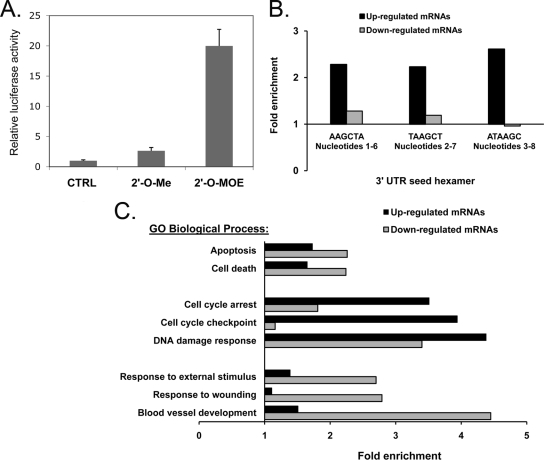
To identify molecular pathways and gain insights into direct mRNA targets affected by miR-21, we suppressed miR-21 in human glioma A172 cells by transfecting them with modified ASOs. We then used Merck Custom human (Agilent) 44K expression arrays to determine mRNA profiles and identify genes whose expression levels were persistently and reproducibly changed by suppression of this miRNA (see Table S1 in the supplemental material). The analysis was performed on RNA samples harvested at 6 and 24 hours posttransfection, consistent with the time frames commonly used for this type of analysis (18, 36, 37). Since modified oligonucleotides can induce a wide range of nonspecific, off-target-mediated effects, several ASO molecules of different chemistries have been used for miR-21 inhibition. To assess the degree of inhibition, we transfected into glioma cells the luciferase reporter of miR-21 activity, a vector that contained a perfect miR-21 binding site downstream of the luciferase gene (pMIR-Report; Ambion) and was therefore utilized for testing miR-21 activity and the efficacy of its inhibitors. Among the molecules tested, the 2′-O-MOE ASO was the most potent in repressing miR-21 activity (Fig. 2A), in agreement with previous data (10), whereas the uniform 2′-O-methyl ASO was the least potent, and mixed locked nucleic acid (LNA)-DNA oligonucleotides were variable (data not shown). In addition to being the most potent ASO, the 2′-O-MOE ASO also induced expression of an mRNA population significantly enriched in miR-21 seeds. In fact, three hexamers, corresponding to nucleotides 1 to 6, 2 to 7, and 3 to 8 of miR-21, were strongly overrepresented in the population of upregulated transcripts (with expectations of 1.45e−09, 1.37e−14, and 1.82e−12, respectively) but not in the population of downregulated transcripts (Fig. 2B). Such overrepresentation of the hexamers suggests an abundance of direct miR-21 targets among the genes upregulated by 2′-O-MOE anti-miR-21 and indicates that the effects of this molecule on glioma cells are indeed mediated through the miR-21 pathway. Other tested inhibitors did not show such a strong bias for regulating miR-21 seed-containing mRNAs (data not shown). We therefore used the 2′-O-MOE anti-miR-21 molecule and the corresponding mismatched control oligonucleotide in subsequent experiments.
FIG. 2.
Analysis of genes regulated by anti-miR-21. (A) Validation of miR-21 inhibitor. Glioma A172 cells were cotransfected with 50 nM miR-21 synthetic antisense inhibitors (CTRL, control oligonucleotide; 2′-O-Me, 2′-O-methyl) and the luciferase sensor of miR-21 activity containing a perfect miR-21 binding site. Anti-miR-21 activity was then measured using the luciferase reporter assay and normalized to Renilla levels. 2′-MOE ASO inhibits miR-21 ∼20-fold. (B) Microarray expression profiling confirms upregulation of endogenous targets. The hexamer composition of the 3′ UTRs of transcripts up- or downregulated (P < 0.05) by anti-miR-21 at 24 h in two replicate experiments was compared to the hexamer composition of the 3′ UTRs of all transcripts on the microarray. The levels of enrichment of miR-21 seed hexamers in the regulated transcripts are shown. (C) Biofunctions of genes regulated by anti-miR-21 identified by microarray profiling. Transcripts up- or downregulated (P < 0.05) by anti-miR-21 in two replicate experiments were mapped to Entrez genes annotated with Gene Ontology (GO) database biological process terms. GO terms were ranked by the hypergeometric P values for their enrichment in the regulated genes compared to the levels for all transcripts in the microarray. The levels of enrichment of the most significantly enriched distinct terms from either up- or downregulated signatures are shown.
Transcriptional profiling of knocked-down miR-21 cells revealed changes in the expression levels of ∼570 genes (P < 0.05) associated with various biological functions (Fig. 2C). DNA damage response genes, regulators of cell cycle arrest, and positive regulators of apoptosis have been enriched among genes upregulated at 24 h. Among downregulated genes, those involved in stress response, apoptosis, and regulation of signal transduction (particularly Jun N-terminal protein kinase cascade, mitogen-activated protein kinase kinase kinase cascade, and stress-activated protein kinase pathway) and, most significantly, genes associated with blood vessel morphogenesis and development have been strongly enriched (expectation value < 10−4) (Fig. 2C). This molecular profiling suggested that miR-21 regulates multiple genes involved in several cellular programs in glioma cells.
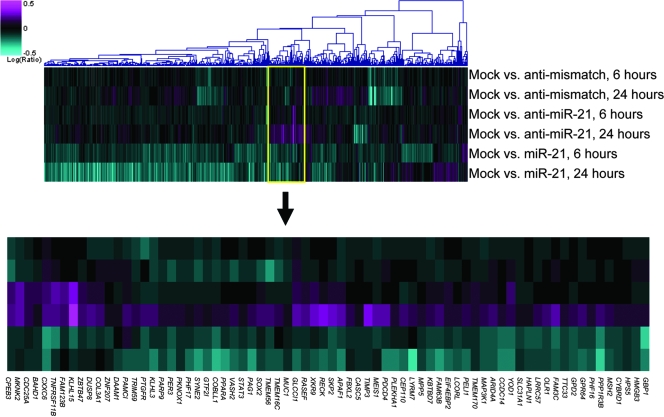
Concurrent with miR-21 inhibition, we have also tested the effects of miR-21 overexpression by transfecting synthetic miR-21 duplex molecules into the A172 glioma cells (Fig. 3). Since, as demonstrated earlier, the endogenous miR-21 is high in gliomas and many other cancer cell types (5, 54), we hypothesized that its overexpression might have additional regulatory effects on miR-21 targets. Notably, a large group of miR-21 octamer- and/or heptamer-containing mRNAs (at least 65) that were upregulated by anti-miR-21 (P < 0.05) has been also downregulated by miR-21 overexpression (P < 0.05) (Fig. 3). Such reverse correlation between modulated miR-21 level and expression levels of this set of mRNAs suggests that this mRNA population is enriched in direct miR-21 targets. An abundance of targets predicted by several commonly used computational algorithms supports this conclusion. Particularly, PDCD4, RECK, SOX2, and PELI1 have been predicted by TargetScan, PicTar, and Miranda (16, 17, 29, 34), and Yod1, PPARA, GPR64, RASGRP1, FAM63B, TIMP3, CDC25A, GLCCI1, TRIM59, CCDC14, PLEKHA1, CPEB3, MSH2, TNFRSF11B, ANKRD46, Sesn1, FAM3c, STAT3, and APAF1 have been predicted either by TargetScan, PicTar, and Miranda or by RNA Hybrid (25, 29, 34, 49). Several of these genes are directly associated with apoptosis. Prominent among them are apoptotic peptidase activating factor 1 (APAF1), a cytoplasmic protein that initiates apoptosis by forming apoptosome (26); PDCD4, a tumor suppressor recently validated as a direct miR-21 target (1, 14), whose expression is often lost in gliomas (15); and tumor necrosis factor receptor superfamily member 11B (TNFRSF11B), which was shown to have a role in different cancers (20).
FIG. 3.
Microarray expression profiling identifies genes regulated by both miR-21 and anti-miR-21. Expression profiles of cells transfected with the control mismatched anti-miR-21 oligonucleotide, the anti-miR-21 oligonucleotide, or the miR-21 mimetic duplex were compared. The top panel is a heat map representation of genes regulated in any one experiment and containing motifs matching the octamer (ATAAGCTA) and heptamer(s) (ATAAGCT and/or TAAGCTA) of miR-21 nucleotides 1 to 8. The bottom panel is a detail of the heat map, showing the cluster of seed-matched genes both upregulated by anti-miR-21 and downregulated by the miR-21 duplex.
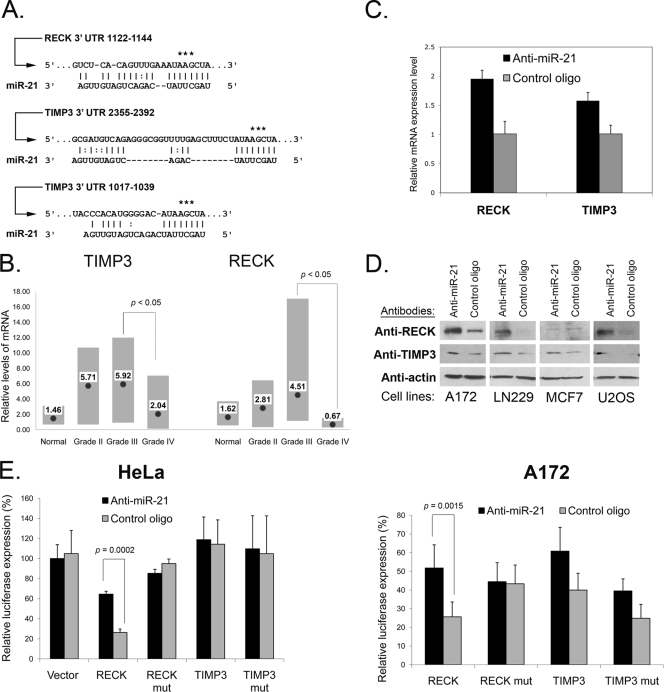
In addition to apoptosis- and cell cycle-related genes, analysis of miR-21 array data identified a number of important ECM remodeling factors that are computationally predicted miR-21 targets (Fig. 3; also see Table S1 in the supplemental material). Remarkably, the mRNA levels of two major inhibitors of MMPs, RECK and TIMP3, have been affected by both miR-21 knockdown and its overexpression. MMPs are a group of peptidases involved in degradation of the ECM. Accumulated data suggested that MMP levels and activities are significantly elevated in human gliomas, which contributes to glioma cell invasion of the surrounding normal tissues, metastasis, and angiogenesis through cell surface ECM degradation (21, 45). mRNAs carrying RECK and TIMP3 are predicted conserved miR-21 targets with one and two putative binding sites, respectively (Fig. 4A). RECK is a membrane-anchored MMP inhibitor whose reduced expression or inactivation seems to be critical for the invasiveness and metastasis of various cancers, including glioma cancer (7, 46). Its expression level is also an important prognostic factor for multiple cancer types (6). However, molecular mechanisms that downregulate RECK level are not well studied. TIMP3 is a tissue inhibitor of MMPs that inhibits angiogenesis and tumor cell infiltration and also induces apoptosis (2, 48). It has been demonstrated to inhibit MMP activity and tumor growth in malignant gliomas in vivo (31, 32). Our mRNA array analysis suggested that RECK and TIMP3 mRNAs may be regulated by miR-21.
FIG. 4.
miR-21 targets RECK and TIMP3. (A) Predicted miR-21 binding sites within RECK and TIMP3 3′ UTRs. The asterisks depict nucleotides mutated for the luciferase reporter assays. (B) RECK and TIMP3 mRNA expression in normal brain tissue and gliomas of different grades was analyzed by qRT-PCR and normalized to GAPDH mRNA levels. Per group, four control brains and grade II gliomas and eight grade III and grade IV gliomas were analyzed. The gray bars represent the data ranges. Mean expression levels are depicted by black dots and corresponding numbers. Since a few samples were analyzed in “normal” and “grade II” sets, the analysis reaches significance (P < 0.05) for the transition from grade III to IV only. (C) RECK and TIMP3 mRNAs are upregulated upon treatment of glioma cells with anti-miR-21. A172 cells were transfected with either the anti-miR-21 or the control oligonucleotide (control oligo) and grown for 48 h, mRNA was isolated, and qRT-PCR for RECK and TIMP3 was performed. The data were normalized to GAPDH mRNA levels. Data are represented as means ± SEM. (D) Western blot validation of RECK and TIMP3 protein upregulation by anti-miR-21 in glioma (A172 and LN229) and other (MCF7, breast carcinoma; U2OS, osteosarcoma) cell lines. The analysis was performed at 72 h posttransfection. (E) pMir-Report vectors containing the wild-type RECK 3′ UTR (RECK wt), RECK 3′ UTR with a mutated miR-21 binding site (RECK mut), the wild-type TIMP3 3′ UTR (TIMP3 wt), or the TIMP3 3′ UTR with both predicted miR-21 binding sites mutated (TIMP3 mut) were cotransfected with either miR-21 inhibitor (Anti-miR-21) or the mismatched control oligonucleotide into A172 and HeLa cells. The inhibition of miR-21 by the antisense inhibitors resulted in a significant increase in luciferase signals of RECK wt- but not RECK mut-transfected cells. No consistent effect of miR-21 inhibition on the TIMP3 3′ UTR was observed. Depicted are the averages of results from representative experiments performed in triplicate. Data are represented as means ± SEM.
miR-21 targets MMP inhibitors.
In order to test our hypothesis, we examined RECK and TIMP3 mRNA levels in the progression of human gliomas (normal brain tissue versus gliomas of different grades) by qRT-PCR. RECK mRNA expression was significantly lower in GBMs than in normal brain tissue and, especially, in grade II and III gliomas. TIMP3 mRNA expression also tended to be lower in GBM tumors than in grade II and III gliomas (Fig. 4B). These data suggest that RECK and TIMP3 may represent natural miR-21 targets in vivo. To validate miR-21 regulation of RECK and TIMP3, we first confirmed RECK and TIMP3 mRNA elevation in miR-21 knocked-down A172 cells by qRT-PCR (Fig. 4C). RECK mRNA level increased consistently ∼2-fold and TIMP3 mRNA about 1.6-fold in the cells transfected with anti-miR-21. We then examined the protein levels of RECK and TIMP3 and found that both proteins were upregulated by miR-21 inhibition in glioma A172 and LN229 cells (Fig. 4D). In glioma U87 cells, TIMP3 was also upregulated by anti-miR-21; however, we could not detect distinct RECK bands by Western blot analyses. Nevertheless, using qRT-PCR, we observed a twofold upregulation of RECK mRNA expression by anti-miR-21 (data not shown), similar to the effect demonstrated for the A172 glioma cell line (Fig. 4C). To validate that miR-21 can bind directly to and regulate RECK and TIMP3 mRNAs through the predicted binding sites, we constructed luciferase reporter plasmids that contain RECK and TIMP3 3′ UTR sequences downstream of firefly luciferase and tested the luciferase activity in HeLa and A172 glioma cells, both expressing high levels of endogenous miR-21. We also constructed mutant versions of these reporters, altering bases in the putative miR-21 binding sites in a way that was predicted to abolish miR-21 binding. All constructs were transfected into cultured HeLa and A172 cells, along with a Renilla luciferase transfection control, and both firefly and Renilla luciferase levels were assayed by luminometry. These experiments have been performed with the miR-21 inhibitor and with a mismatched control oligonucleotide to ensure that any observed effects are driven by miR-21. These results indicate that RECK is a direct miR-21 target (Fig. 4E). The TIMP3 3′ UTR construct did not consistently respond to changes in miR-21 levels, indicating that TIMP3 is not likely to be a direct miR-21 target (Fig. 4E). However, since TIMP3 mRNA and protein levels were modulated by miR-21 expression (Fig. 4C and D), TIMP3 is likely to be a downstream effector of miR-21 signaling.
Inhibition of miR-21 leads to downregulation of MMP activities.
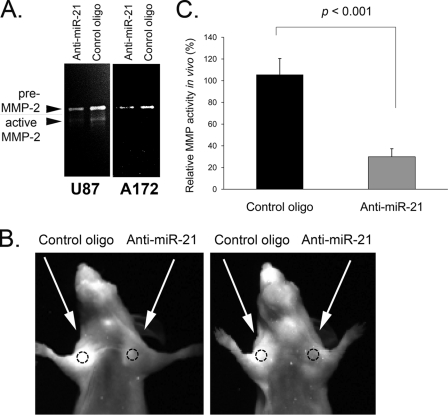
Since the expression levels of at least two MMP inhibitors, RECK and TIMP3, are regulated in glioma cells by miR-21, we next examined the effects of miR-21 inhibition on MMP proteolytic activities. Conditioned media from glioma A172 and U87 cells transfected with either miR-21 inhibitor or the control oligonucleotide were analyzed by gelatin zymography. In this assay, major bands that correspond to the active form of MMP-2 and/or pre-MMP-2 were detected in glioma cell lines. When miR-21 was suppressed, the amount of gelatin digested that reflects the levels of MMP-2 proteolytic activity was significantly reduced relative to the amount for the cells transfected with the control oligonucleotide (Fig. 5A). Notably, total protein levels of secreted MMP-2 were not affected by miR-21 suppression (data not shown). Concurring with previously published reports (12, 46), our data indicate that these MMP inhibitors regulate MMP activity posttranslationally.
FIG. 5.
miR-21 inhibition affects MMP activity in vitro and in vivo. (A) miR-21 inhibition reduces the activity of gelatinolytic enzymes in glioma A172 and U87 cells. Fresh media of cell cultures transfected with either anti-miR-21 or the corresponding control oligonucleotide were analyzed by gelatin zymography. (B) In vivo imaging of MMP activity. Nude mice were injected subcutaneously in the flank with U87 cells transfected with 50 nM anti-miR-21 (left side) or control oligonucleotides (right side). At 3 days postinjection, the MMP activity was measured using MMPsense (Visen) and fluorescence imaging. Shown are two representative mice of a group of eight mice. (C) Quantitation of the relative fluorescence intensities shown in panel B by use of ImageJ software (NIH). Quantitation is demonstrated for a representative experiment (n = 4). The experiment was repeated three times. Data are represented as means ± SEM.
We next studied the effects of miR-21 inhibition on MMP proteolytic function during glioma growth in vivo. In these experiments, we used tumorigenic U87 cells, a common model of human gliomas (23). U87 cells transfected with either anti-miR-21 or the control oligonucleotide were implanted into nude mice subcutaneously, and MMP activities were assessed by using an MMPSense 680 imaging probe (VisEn Medical). This probe is a protease-activated fluorescent agent, optically silent in its inactive state and highly fluorescent following MMP-mediated activation. It allows in vivo imaging of key MMPs, including MMP-2 as well as MMP-3, -9, -7, -12, and -13 (4, 32). Each mouse was implanted subcutaneously with anti-miR-21-transfected U87 cells on one side of the body and control U87 cells on the other side. At 3 days after cell implantation, MMPsense was injected intravenously, and the mice were visualized using a fluorescence imaging system 24 h later. All tumors produced from miR-21-inhibited cells demonstrated significantly lower MMP activity than control tumors (Fig. 5B and C). The relative fluorescence intensities were determined using ImageJ software (NIH), and MMP signals were normalized to tumor size, measured by a caliper. The anti-miR-21 oligonucleotide used in this study did not affect subcutaneous U87 tumor growth within the first week after cell implantation, when the imaging was performed (data not shown).
Inhibition of miR-21 reduces motility and invasiveness of glioma cells.
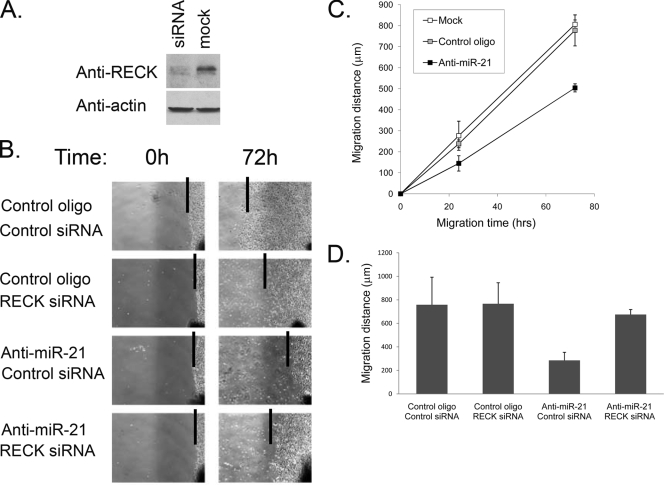
MMPs can degrade ECM components and therefore play important roles in mediating GBM tumor cell motility and invasion (45). Moreover, MMP-2 and MMP-9 activities correlate well with glioma cell migration and invasiveness (58). We have therefore tested miR-21 effects on glioma cell motility and invasion. First, we performed an in vitro scratch motility assay on A172 cells (35). Cells were plated, allowed to grow to confluence, and transfected with anti-miR-21 or the mismatched control oligonucleotide. The monolayer was then scratched to create a cleared area within the monolayer. Pictures were taken, and the two-dimensional movement of the transfected cells was quantified by measuring the migration distance at different time points and comparing it with the front area at time zero. These experiments demonstrated that inhibition of miR-21 reduced the motility of glioma cells (Fig. 6B and C). Notably, this reduced-motility phenotype was partly rescued by cotransfection of anti-miR-21 with small interfering RNA (siRNA) to RECK (Fig. 6B and D), indicating that miR-21 regulation of glioma cell motility is mediated, at least partly, by RECK.
FIG. 6.
Inhibition of miR-21 affects glioma cell motility. (A) Validation of RECK downregulation by siRNA. A172 cells were transfected with RECK siRNA or Lipofectamine only (mock). At 72 h following transfection, cells were harvested and subjected to Western blot analysis using anti-RECK antibodies. (B) Confluent A172 cells were transfected with either miR-21 inhibitor or the control oligonucleotide in combination with RECK siRNA or control siRNA. At 5 h posttransfection, the cell monolayer was scratched with a scratch spatula (time zero) and migration of the cells toward the “wound” was visualized. Images were taken at 0 and 72 h after the monolayer was scratched. The software program MetaVue was used to determine the migration distance. Depicted are representative images and indications of the migration distances (lines at monolayer fronts). (C) Confluent A172 cells were transfected in triplicate with the miR-21 inhibitor, the mismatched control oligonucleotide, or no oligonucleotide (mock). The cell monolayers were scratched, and migration of the cells toward the “wound” was visualized as described for panel B. Images were taken at various time points after the monolayer was scratched, and the software program MetaVue was used to determine the migration distance. The experiment was repeated three times. Depicted are the average values of results from a representative experiment. Data are represented as means ± SEM. (D) Quantification of the experiment whose results are shown in panel B. Images were taken at various time points after the monolayer was scratched, and the software program MetaVue was used to determine the migration distance. Inhibition of miR-21 resulted in a significant reduction in the migration distance of the monolayer front (P < 0.01), which was rescued by cotransfection with RECK siRNA. Depicted are the averages of results from representative experiments performed in triplicate. Data are represented as means ± SEM.
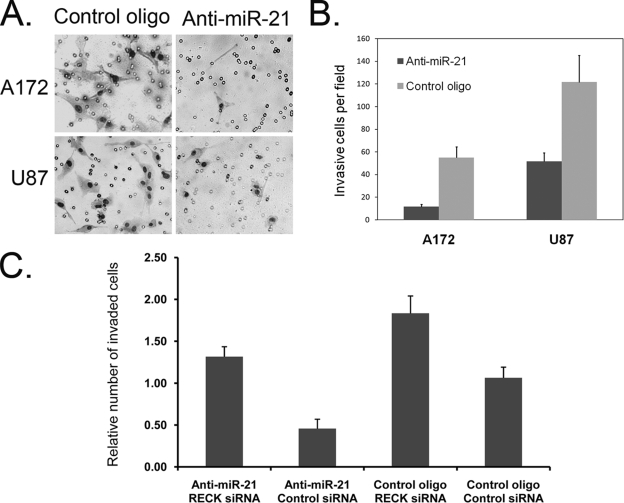
To measure miR-21 effects on glioma cell invasiveness, a better indication of glioma migratory and invasive properties in vivo, we employed a Transwell invasion system. The system consists of two fluid-filled, stacked compartments separated by a porous membrane filter coated with Matrigel. Cells were grown in the upper chamber and assessed for invasion through the Matrigel toward a chemoattractant (10% serum) in the lower chamber. The numbers of invasive A172 and U87 cells in cultures with inhibited miR-21 were significantly reduced relative to those in cultures transfected with the control oligonucleotide (Fig. 7A and B). This reduced invasiveness was not observed, however, when anti-miR-21 was cotransfected with RECK siRNA (Fig. 7C), suggesting that RECK is the major factor that mediates miR-21 function in glioma invasion, at least in vitro.
FIG. 7.
Inhibition of miR-21 reduces glioma cell invasiveness. The Matrigel invasion assay was performed on glioma cell lines (A172 and U87) transfected with either the inhibitor of miR-21 (anti-miR-21) or the control oligonucleotide (control oligo). Invaded cells were fixed, stained, and observed by optical microscopy. The experiment was performed in triplicate and repeated three times. (A) Pictures of representative cell fields for each treatment were taken by a camera connected to the microscope with ×10 magnification. (B) Numerical representation of the data obtained by counting average numbers of cells from three different fields for each treatment. Data are represented as means ± SEM. (C) A172 cells were cotransfected with the indicated oligonucleotides (anti-miR-21, RECK siRNA, or the corresponding control 2′-O-MOE and siRNA oligonucleotides). RECK siRNA increased the invasiveness and rescued the effect of anti-miR-21. Data are represented as means ± SEM.
DISCUSSION
miR-21 is one of the most commonly implicated miRNAs in cancer. Its expression is highly upregulated in a variety of solid tumors, including GBM, breast, lung, colon, prostate, pancreas, and stomach cancers (5, 54). Elevated miR-21 expression has been causally linked to proliferation, apoptosis, and migration of several cancer cell lines (1, 5, 44, 60). However, our knowledge of the molecular mechanisms mediating miR-21 function in cancer generally and gliomas specifically is limited. According to recent findings, several factors can upregulate miR-21 expression. Among them are signal transducer and activator of transcription 3 (STAT3) (39), which is constitutively activated in a variety of cancer types, including malignant gliomas. The miR-21 gene contains two conserved STAT3 binding sites within its enhancer region and is induced by interleukin 6 (IL-6) in a STAT3-dependent way. Another factor that can induce miR-21 expression is hypoxia, a general feature of all solid tumors (30). In GBM/glioma progression, both STAT3 and Hif1a, a major mediator of cell response to hypoxia, are upregulated (47, 52) and can possibly account for miR-21 overexpression.
mRNA expression profiling for revealing miR-21 functions.
In this study, we attempted to identify major mRNA targets and signaling pathways that mediate miR-21 regulation in gliomas. We used both gain- and loss-of-function approaches and combined them with 44K Merck Custom human expression array analysis to study miR-21 signaling in glioma cells (Fig. 2B and C and 3; also see Table S1 in the supplemental material). Such a bidirectional approach to manipulating miR-21 levels enabled narrowing of the lists of regulated genes and identification of those whose expression correlated negatively with miR-21 levels, e.g., upregulated under miR-21 inhibition conditions and downregulated by its overexpression. Though mRNA expression analysis does not allow identification of targets whose translation is regulated without modulation of mRNA levels, the advantages of such an approach are multifold. First, it enables bioinformatic analysis for determination of biological functions and signaling pathways affected and therefore can predict functions that a particular miRNA may play. Second, it allows sequence analysis and identification of genes showing predicted miRNA-binding motifs and enrichment in seeds (putative direct targets). Importantly, since both inhibition and overexpression of an miRNA with synthetic oligonucleotides can potentially cause significant off-target effects, enrichment of the predicted targets among negatively regulated genes validates the specificity of the response and enables an intelligent design of most specific miRNA inhibitors and mimics. Here, we show that a 2′-O-MOE ASO not only is potent but also upregulates miR-21 seed-containing mRNAs (Fig. 2 and 3).
Inhibition of miR-21 leads to a decrease of MMP activities.
Our mRNA expression analysis suggests that miR-21 regulates the expression of multiple mRNA targets and thereby induces several cellular programs in glioma cells, including those associated with the tumor invasiveness and microvascular proliferation typical for the GBM as well as those involved in cell cycle regulation and apoptosis. Many genes identified in this study are potent signal transducers and regulators of gene expression implicated in glioma biology and more generally in carcinogenesis. In this study, we have demonstrated in vitro and in vivo, using U87 glioma xenografts in nude mice, that miR-21 regulates MMPs and glioma cell invasiveness by directly controlling the MMP inhibitor RECK. RECK, a membrane-anchored regulator, and TIMP3, the ECM-bound protease regulator, are key inhibitors of several MMPs, and their expression is prognostic in a number of common cancers (28, 53). miR-21 negatively regulates the mRNA and protein levels of RECK and TIMP3 in cultured glioma cells (Fig. 4C and D). Inhibition of miR-21 decreases MMP activities both in vitro and in vivo (Fig. 5). Moreover, it leads to reduction of glioma cell motility and invasion, which is mediated by elevated RECK expression and was partly rescued by RECK siRNA (Fig. 6 and 7). In glioma progression from less invasive grade II gliomas to very invasive GBMs, RECK and TIMP3 levels drop (Fig. 4B), whereas miR-21 expression increases (Fig. 1). This implies that miR-21 regulates RECK and TIMP3 expression and thus MMP activities and glioma tumor invasiveness.
In addition to MMP inhibition, TIMP3 may contribute to blocking of cancer progression by participating in a number of cellular processes, including apoptosis. Its ectopic expression in glioma cells induces upregulation of genes associated with angiogenesis, growth factors, cytokines, death receptors, and substrates of the various MMPs (31). Importantly, it inhibits tumor necrosis factor alpha-converting enzyme and induces apoptosis through the stabilization of tumor necrosis factor alpha receptors on the cell surface (42). TIMP3 has been demonstrated to activate caspases and therefore induce apoptosis of human glioma cells in vitro and the growth inhibition of human glioma tumors in a mouse model (31, 32).
Biological functions of other miR-21 targets.
In addition to RECK and TIMP3, many genes respond to both increased and decreased levels of miR-21 (Fig. 3; also see Table S1 in the supplemental material), including several that play important functions in glioma biology and in carcinogenesis. For example, APAF1 (apoptotic protease activating factor 1) is the molecular core of apoptosome. It is typically required for activation of the caspases that initiate apoptosis (3, 26). The APAF1 3′ UTR contains a strong miR-21 binding site (9-mer binding at miR-21 5′ end), and therefore, it is likely one of the direct miR-21 targets. In gliomas, APAF1 is often inactivated or downregulated (56). Our data suggest that this can be at least partly due to miR-21 regulation, in addition to the reported chromosome 12q22-23 loss of heterozygosity and hypermethylation (56). Its overexpression by viral transduction induces apoptosis in glioma cells and may be beneficial in glioma treatment (50). STAT3, the other gene that may play a tumor suppressor function in GBM (11), is also negatively regulated by miR-21 according to the microarray data (see Table S1 in the supplemental material) and is a predicted miR-21 target (29, 34). Interestingly, STAT3 can potentially mediate IL-6-miR-21 autocrine feedback. As mentioned, both STAT3 and IL-6 are usually elevated in malignant gliomas (57) and can be among the factors inducing miR-21 expression (39). miR-21, in turn, may downregulate STAT3 (see Table S1 in the supplemental material). Such a regulatory loop between miR-21 and IL-6/STAT3 may provide a feedback mechanism for stabilizing miR-21 expression and balancing STAT3 signaling.
Several mRNAs regulated by miR-21 and anti-miR-21 in glioma cells (Fig. 3; also see Table S1 in the supplemental material) have recently been identified in an mRNA expression analysis of miR-21 knocked-down MCF7 breast carcinoma cells (14). Among these mRNAs are PDCD4, APAF1, FAM3C, SESN1, and GLCCI1. Our analysis also confirmed tropomyosin 1 (TPM1) regulation by miR-21, previously reported to occur in MCF7 cells (59, 60). As identified in our study, TIMP3 or both TIMP3 and RECK are also induced by anti-miR-21 in MCF7 (breast carcinoma) and U2OS (osteosarcoma) cells (Fig. 4D). However, we have not detected any correlation between miR-21-modulated expression in several glioma cell lines and protein level of phosphatase and tensin homolog (data not shown), the reported miR-21 target in hepatocellular carcinomas (44). These combined results indicate that miR-21 regulates overlapping though not identical sets of genes in different cells and therefore shares common signaling pathways in different cancers.
Issues of miRNA inhibitors and glioma models.
We note that, though 2′-O-MOE miR-21 ASO induced many proapoptotic and cell cycle arrest genes (Fig. 2C; also see Table S1 in the supplemental material), it did not significantly and consistently change the viability of glioma cell lines, which was reported for other, less potent miR-21 inhibitors (5, 8). This strong and specific 2′-O-MOE anti-miR-21 ASO also did not cause a reduction of subcutaneous tumor growth in a U87 glioma model (data not shown). In contrast, an LNA-DNA anti-miR-21 oligonucleotide was previously shown to reduce tumors in an intracranial U87 glioma model (8). Although the locations of the tumors in the mice were different, not fully allowing a direct comparison, the likely explanation for these conflicting results might be that the suboptimally designed LNA-DNA oligonucleotide used in the earlier study killed glioma cells by an off-target mechanism. We should also note that subcutaneous U87 tumors are noninfiltrative, noninvasive, and poorly vascularized. Therefore, though commonly used and very robust, they do not represent an accurate model for studying invasiveness and its effects on tumor growth of human GBMs. Better models will be developed and employed in the future to study the effects of miR-21 inhibition on glioma growth and invasion.
Considered together, this study and previous reports show that differences in nucleic acid chemistry can, in some cases, profoundly alter cellular responses to miRNA inhibition. More-careful analyses that combine mRNA expression profiling with studies of inhibitor potency should help to ensure that cellular responses to miRNA inhibition are specific and genuine. While the goal of this study was to better understand miR-21 function in gliomas, future research is required to address the therapeutic potential of modulating miR-21. Our data suggest that by modulating expression and/or activity of one miRNA with specific tools, e.g., inhibitors or mimics, it is possible to control the expression of multiple oncogenic or tumor-suppressing genes and simultaneously trigger several cellular programs that these genes mediate.
Supplementary Material
Acknowledgments
We thank Xandra O. Breakefield for helpful discussions and use of her laboratory facilities (GTBT CA6924) and Ralph Weissleder for providing the MMP probe and access to the Imaging Facility (NIH-NCI P50 CA86355-04). We thank John Pham for significant intellectual and experimental contribution to this work. A plasmid coding for the luciferase reporter of miR-21 activity was a kind gift from Ambion.
This study was supported by NCI R21CA116141 and Brain Tumor Society grants (to A.M.K.), a Paul Brazen American Brain Tumor Association Fellowship (to G.G.), and a Steve Kaplan American Brain Tumor Association Fellowship (to T.W.).
Footnotes
Published ahead of print on 30 June 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Asangani, I. A., S. A. Rasheed, D. A. Nikolova, J. H. Leupold, N. H. Colburn, S. Post, and H. Allgayer. 2008. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 272128-2136. [DOI] [PubMed] [Google Scholar]
- 2.Baker, A. H., S. J. George, A. B. Zaltsman, G. Murphy, and A. C. Newby. 1999. Inhibition of invasion and induction of apoptotic cell death of cancer cell lines by overexpression of TIMP-3. Br. J. Cancer 791347-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bratton, S. B., G. Walker, S. M. Srinivasula, X. M. Sun, M. Butterworth, E. S. Alnemri, and G. M. Cohen. 2001. Recruitment, activation and retention of caspases-9 and -3 by Apaf-1 apoptosome and associated XIAP complexes. EMBO J. 20998-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bremer, C., C. H. Tung, and R. Weissleder. 2001. In vivo molecular target assessment of matrix metalloproteinase inhibition. Nat. Med. 7743-748. [DOI] [PubMed] [Google Scholar]
- 5.Chan, J. A., A. M. Krichevsky, and K. S. Kosik. 2005. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 656029-6033. [DOI] [PubMed] [Google Scholar]
- 6.Clark, J. C., D. M. Thomas, P. F. Choong, and C. R. Dass. 2007. RECK—a newly discovered inhibitor of metastasis with prognostic significance in multiple forms of cancer. Cancer Metastasis Rev. 26675-683. [DOI] [PubMed] [Google Scholar]
- 7.Correa, T. C., C. A. Brohem, S. M. Winnischofer, L. B. da Silva Cardeal, R. M. Sasahara, S. R. Taboga, M. C. Sogayar, and S. S. Maria-Engler. 2006. Downregulation of the RECK-tumor and metastasis suppressor gene in glioma invasiveness. J. Cell. Biochem. 99156-167. [DOI] [PubMed] [Google Scholar]
- 8.Corsten, M. F., R. Miranda, R. Kasmieh, A. M. Krichevsky, R. Weissleder, and K. Shah. 2007. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 678994-9000. [DOI] [PubMed] [Google Scholar]
- 9.Croce, C. M. 2008. Oncogenes and cancer. N. Engl. J. Med. 358502-511. [DOI] [PubMed] [Google Scholar]
- 10.Davis, S., B. Lollo, S. Freier, and C. Esau. 2006. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 342294-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Iglesia, N., G. Konopka, S. V. Puram, J. A. Chan, R. M. Bachoo, M. J. You, D. E. Levy, R. A. Depinho, and A. Bonni. 2008. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 22449-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.English, J. L., Z. Kassiri, I. Koskivirta, S. J. Atkinson, M. Di Grappa, P. D. Soloway, H. Nagase, E. Vuorio, G. Murphy, and R. Khokha. 2006. Individual Timp deficiencies differentially impact pro-MMP-2 activation. J. Biol. Chem. 28110337-10346. [DOI] [PubMed] [Google Scholar]
- 13.Esquela-Kerscher, A., and F. J. Slack. 2006. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer. 6259-269. [DOI] [PubMed] [Google Scholar]
- 14.Frankel, L. B., N. R. Christoffersen, A. Jacobsen, M. Lindow, A. Krogh, and A. H. Lund. 2008. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2831026-1033. [DOI] [PubMed] [Google Scholar]
- 15.Gao, F., P. Zhang, C. Zhou, J. Li, Q. Wang, F. Zhu, C. Ma, W. Sun, and L. Zhang. 2007. Frequent loss of PDCD4 expression in human glioma: possible role in the tumorigenesis of glioma. Oncol. Rep. 17123-128. [PubMed] [Google Scholar]
- 16.Griffiths-Jones, S., R. J. Grocock, S. van Dongen, A. Bateman, and A. J. Enright. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34D140-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimson, A., K. K. Farh, W. K. Johnston, P. Garrett-Engele, L. P. Lim, and D. P. Bartel. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 2791-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, L., X. He, L. P. Lim, E. de Stanchina, Z. Xuan, Y. Liang, W. Xue, L. Zender, J. Magnus, D. Ridzon, A. L. Jackson, P. S. Linsley, C. Chen, S. W. Lowe, M. A. Cleary, and G. J. Hannon. 2007. A microRNA component of the p53 tumour suppressor network. Nature 4471130-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, L., X. He, S. W. Lowe, and G. J. Hannon. 2007. microRNAs join the p53 network—another piece in the tumour-suppression puzzle. Nat. Rev. Cancer. 7819-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holen, I., and C. M. Shipman. 2006. Role of osteoprotegerin (OPG) in cancer. Clin. Sci. (London) 110279-291. [DOI] [PubMed] [Google Scholar]
- 21.Hu, B., P. Guo, Q. Fang, H. Q. Tao, D. Wang, M. Nagane, H. J. Huang, Y. Gunji, R. Nishikawa, K. Alitalo, W. K. Cavenee, and S. Y. Cheng. 2003. Angiopoietin-2 induces human glioma invasion through the activation of matrix metalloprotease-2. Proc. Natl. Acad. Sci. USA 1008904-8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irizarry, R. A., B. M. Bolstad, F. Collin, L. M. Cope, B. Hobbs, and T. P. Speed. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishii, N., D. Maier, A. Merlo, M. Tada, Y. Sawamura, A. C. Diserens, and E. G. Van Meir. 1999. Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol. 9469-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson, A. L., S. R. Bartz, J. Schelter, S. V. Kobayashi, J. Burchard, M. Mao, B. Li, G. Cavet, and P. S. Linsley. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21635-637. [DOI] [PubMed] [Google Scholar]
- 25.John, B., A. J. Enright, A. Aravin, T. Tuschl, C. Sander, and D. S. Marks. 2004. Human microRNA targets. PLoS Biol. 2e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, C. E., Y. Y. Huang, A. B. Parrish, M. I. Smith, A. E. Vaughn, Q. Zhang, K. M. Wright, T. Van Dyke, R. J. Wechsler-Reya, S. Kornbluth, and M. Deshmukh. 2007. Differential Apaf-1 levels allow cytochrome c to induce apoptosis in brain tumors but not in normal neural tissues. Proc. Natl. Acad. Sci. USA 10420820-20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent, O. A., and J. T. Mendell. 2006. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene 256188-6196. [DOI] [PubMed] [Google Scholar]
- 28.Kotzsch, M., J. Farthmann, A. Meye, S. Fuessel, G. Baretton, V. C. Tjan-Heijnen, M. Schmitt, T. Luther, F. C. Sweep, V. Magdolen, and P. N. Span. 2005. Prognostic relevance of uPAR-del4/5 and TIMP-3 mRNA expression levels in breast cancer. Eur. J. Cancer. 412760-2768. [DOI] [PubMed] [Google Scholar]
- 29.Krek, A., D. Grun, M. N. Poy, R. Wolf, L. Rosenberg, E. J. Epstein, P. MacMenamin, I. da Piedade, K. C. Gunsalus, M. Stoffel, and N. Rajewsky. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37495-500. [DOI] [PubMed] [Google Scholar]
- 30.Kulshreshtha, R., M. Ferracin, S. E. Wojcik, R. Garzon, H. Alder, F. J. Agosto-Perez, R. Davuluri, C. G. Liu, C. M. Croce, M. Negrini, G. A. Calin, and M. Ivan. 2007. A microRNA signature of hypoxia. Mol. Cell. Biol. 271859-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam, P., K. Sian Lim, S. Mei Wang, and K. M. Hui. 2005. A microarray study to characterize the molecular mechanism of TIMP-3-mediated tumor rejection. Mol. Ther. 12144-152. [DOI] [PubMed] [Google Scholar]
- 32.Lamfers, M. L., D. Gianni, C. H. Tung, S. Idema, F. H. Schagen, J. E. Carette, P. H. Quax, V. W. Van Beusechem, W. P. Vandertop, C. M. Dirven, E. A. Chiocca, and W. R. Gerritsen. 2005. Tissue inhibitor of metalloproteinase-3 expression from an oncolytic adenovirus inhibits matrix metalloproteinase activity in vivo without affecting antitumor efficacy in malignant glioma. Cancer Res. 659398-9405. [DOI] [PubMed] [Google Scholar]
- 33.Landgraf, P., M. Rusu, R. Sheridan, A. Sewer, N. Iovino, A. Aravin, S. Pfeffer, A. Rice, A. O. Kamphorst, M. Landthaler, C. Lin, N. D. Socci, L. Hermida, V. Fulci, S. Chiaretti, R. Foa, J. Schliwka, U. Fuchs, A. Novosel, R. U. Muller, B. Schermer, U. Bissels, J. Inman, Q. Phan, M. Chien, D. B. Weir, R. Choksi, G. De Vita, D. Frezzetti, H. I. Trompeter, V. Hornung, G. Teng, G. Hartmann, M. Palkovits, R. Di Lauro, P. Wernet, G. Macino, C. E. Rogler, J. W. Nagle, J. Ju, F. N. Papavasiliou, T. Benzing, P. Lichter, W. Tam, M. J. Brownstein, A. Bosio, A. Borkhardt, J. J. Russo, C. Sander, M. Zavolan, and T. Tuschl. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 1291401-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis, B. P., C. B. Burge, and D. P. Bartel. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 12015-20. [DOI] [PubMed] [Google Scholar]
- 35.Liang, C. C., A. Y. Park, and J. L. Guan. 2007. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2329-333. [DOI] [PubMed] [Google Scholar]
- 36.Lim, L. P., N. C. Lau, P. Garrett-Engele, A. Grimson, J. M. Schelter, J. Castle, D. P. Bartel, P. S. Linsley, and J. M. Johnson. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433769-773. [DOI] [PubMed] [Google Scholar]
- 37.Linsley, P. S., J. Schelter, J. Burchard, M. Kibukawa, M. M. Martin, S. R. Bartz, J. M. Johnson, J. M. Cummins, C. K. Raymond, H. Dai, N. Chau, M. Cleary, A. L. Jackson, M. Carleton, and L. Lim. 2007. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol. Cell. Biol. 272240-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 39.Loffler, D., K. Brocke-Heidrich, G. Pfeifer, C. Stocsits, J. Hackermuller, A. K. Kretzschmar, R. Burger, M. Gramatzki, C. Blumert, K. Bauer, H. Cvijic, A. K. Ullmann, P. F. Stadler, and F. Horn. 2007. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 1101330-1333. [DOI] [PubMed] [Google Scholar]
- 40.Louis, D. N. 2006. Molecular pathology of malignant gliomas. Annu. Rev. Pathol. 197-117. [DOI] [PubMed] [Google Scholar]
- 41.Ma, L., J. Teruya-Feldstein, and R. A. Weinberg. 2007. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449682-688. [DOI] [PubMed] [Google Scholar]
- 42.Mannello, F., and G. Gazzanelli. 2001. Tissue inhibitors of metalloproteinases and programmed cell death: conundrums, controversies and potential implications. Apoptosis 6479-482. [DOI] [PubMed] [Google Scholar]
- 43.Mayr, C., M. T. Hemann, and D. P. Bartel. 2007. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 3151576-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng, F., R. Henson, H. Wehbe-Janek, K. Ghoshal, S. T. Jacob, and T. Patel. 2007. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133647-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakada, M., Y. Okada, and J. Yamashita. 2003. The role of matrix metalloproteinases in glioma invasion. Front. Biosci. 8e261-e269. [DOI] [PubMed] [Google Scholar]
- 46.Oh, J., R. Takahashi, S. Kondo, A. Mizoguchi, E. Adachi, R. M. Sasahara, S. Nishimura, Y. Imamura, H. Kitayama, D. B. Alexander, C. Ide, T. P. Horan, T. Arakawa, H. Yoshida, S. Nishikawa, Y. Itoh, M. Seiki, S. Itohara, C. Takahashi, and M. Noda. 2001. The membrane-anchored MMP inhibitor RECK is a key regulator of extracellular matrix integrity and angiogenesis. Cell 107789-800. [DOI] [PubMed] [Google Scholar]
- 47.Phillips, H. S., S. Kharbanda, R. Chen, W. F. Forrest, R. H. Soriano, T. D. Wu, A. Misra, J. M. Nigro, H. Colman, L. Soroceanu, P. M. Williams, Z. Modrusan, B. G. Feuerstein, and K. Aldape. 2006. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9157-173. [DOI] [PubMed] [Google Scholar]
- 48.Qi, J. H., Q. Ebrahem, N. Moore, G. Murphy, L. Claesson-Welsh, M. Bond, A. Baker, and B. Anand-Apte. 2003. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat. Med. 9407-415. [DOI] [PubMed] [Google Scholar]
- 49.Rehmsmeier, M., P. Steffen, M. Hochsmann, and R. Giegerich. 2004. Fast and effective prediction of microRNA/target duplexes. RNA 101507-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shinoura, N., N. Yamamoto, Y. Yoshida, A. Asai, T. Kirino, and H. Hamada. 2000. Adenovirus-mediated transfer of caspase-8 in combination with superrepressor of NF-kappaB drastically induced apoptosis in gliomas. Biochem. Biophys. Res. Commun. 271544-552. [DOI] [PubMed] [Google Scholar]
- 51.Si, M. L., S. Zhu, H. Wu, Z. Lu, F. Wu, and Y. Y. Mo. 2007. miR-21-mediated tumor growth. Oncogene 262799-2803. [DOI] [PubMed] [Google Scholar]
- 52.Sun, L., A. M. Hui, Q. Su, A. Vortmeyer, Y. Kotliarov, S. Pastorino, A. Passaniti, J. Menon, J. Walling, R. Bailey, M. Rosenblum, T. Mikkelsen, and H. A. Fine. 2006. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 9287-300. [DOI] [PubMed] [Google Scholar]
- 53.Takenaka, K., S. Ishikawa, Y. Kawano, K. Yanagihara, R. Miyahara, Y. Otake, Y. Morioka, C. Takahashi, M. Noda, H. Wada, and F. Tanaka. 2004. Expression of a novel matrix metalloproteinase regulator, RECK, and its clinical significance in resected non-small cell lung cancer. Eur. J. Cancer 401617-1623. [DOI] [PubMed] [Google Scholar]
- 54.Volinia, S., G. A. Calin, C. G. Liu, S. Ambs, A. Cimmino, F. Petrocca, R. Visone, M. Iorio, C. Roldo, M. Ferracin, R. L. Prueitt, N. Yanaihara, G. Lanza, A. Scarpa, A. Vecchione, M. Negrini, C. C. Harris, and C. M. Croce. 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 1032257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, B., T. M. Love, M. E. Call, J. G. Doench, and C. D. Novina. 2006. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol. Cell 22553-560. [DOI] [PubMed] [Google Scholar]
- 56.Watanabe, T., Y. Hirota, Y. Arakawa, H. Fujisawa, O. Tachibana, M. Hasegawa, J. Yamashita, and Y. Hayashi. 2003. Frequent LOH at chromosome 12q22-23 and Apaf-1 inactivation in glioblastoma. Brain Pathol. 13431-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weissenberger, J., S. Loeffler, A. Kappeler, M. Kopf, A. Lukes, T. A. Afanasieva, A. Aguzzi, and J. Weis. 2004. IL-6 is required for glioma development in a mouse model. Oncogene 233308-3316. [DOI] [PubMed] [Google Scholar]
- 58.Wild-Bode, C., M. Weller, and W. Wick. 2001. Molecular determinants of glioma cell migration and invasion. J. Neurosurg. 94978-984. [DOI] [PubMed] [Google Scholar]
- 59.Zhu, S., M. L. Si, H. Wu, and Y. Y. Mo. 2007. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J. Biol. Chem. 28214328-14336. [DOI] [PubMed] [Google Scholar]
- 60.Zhu, S., H. Wu, F. Wu, D. Nie, S. Sheng, and Y. Y. Mo. 2008. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 18350-359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.