Abstract
Methylation is a common modification encountered in DNA, RNA and proteins. It plays a central role in gene expression, protein function and mRNA translation. Prokaryotic and eukaryotic class I translation termination factors are methylated on the glutamine of the essential and universally conserved GGQ motif, in line with an important cellular role. In eukaryotes, this modification is performed by the Mtq2-Trm112 holoenzyme. Trm112 activates not only the Mtq2 catalytic subunit but also two other tRNA methyltransferases (Trm9 and Trm11). To understand the molecular mechanisms underlying methyltransferase activation by Trm112, we have determined the 3D structure of the Mtq2-Trm112 complex and mapped its active site. Using site-directed mutagenesis and in vivo functional experiments, we show that this structure can also serve as a model for the Trm9-Trm112 complex, supporting our hypothesis that Trm112 uses a common strategy to activate these three methyltransferases.
INTRODUCTION
Methylation is a widespread modification occurring on a large variety of substrates. Among these, components linked to protein synthesis (rRNA, tRNA, ribosomal proteins and translational factors) seem to be over-represented (1,2). In most instances, this modification is catalysed by S-adenosylmethionine- (SAM)-dependent methyltransferases (MTases), whose larger family (class I) consists of a seven-stranded β-sheet surrounded by helices on each side (3). The fold of the members of this family is well conserved despite little sequence identity.
Among the various substrates of protein MTases identified so far, the universally conserved GGQ motif found in class I translation termination factors is of particular interest. This motif becomes N5-methylated on the glutamine (Gln) side chain in both prokaryotes and eukaryotes (4,5), although the factors from these two kingdoms are structurally unrelated (6,7). These proteins (RF1 and RF2 in bacteria and eRF1 in eukaryotes) recognize stop codons entering the ribosomal A-site and induce the hydrolysis of the ester bond connecting the newly synthesized polypeptide to the P-site tRNA (8). Hydrolysis is catalysed by the peptidyl transferase centre (PTC) of the large ribosomal subunit and this step requires the entry of the GGQ motif into the PTC (9–11).
In Escherichia coli, the MTase PrmC catalyses the Gln modification (12,13). PrmC comprises two domains: an N-terminal five-helix bundle connected by a short β-hairpin to a large C-terminal domain characteristic of the class I SAM–dependent MTases (14,15). The crystal structure of the complex between E. coli RF1 and PrmC has revealed that this MTase is specific for the closed form of RF1 (14). This methylation is clearly necessary for efficient bacterial translation termination in vivo, since prmC inactivation reduces the specific termination activity of RFs 3- to 4-fold. In E. coli, K12 strains, which carry a mutation reducing RF2 activity, lack of methylation is no longer compatible with normal cell growth. In strains with normal RF2 activity, prmC inactivation considerably reduces growth on poor carbon sources, suggesting that lack of RF methylation limits the synthesis of some proteins that are important under such conditions (16). Similar results were obtained in vitro, where the efficiency of termination with unmethylated RF2 is at least three times lower than with methylated (4). Recent high-resolution crystal structures of 70S ribosome termination complexes bound with RF1 or RF2 and mRNA containing a stop codon have revealed the mechanism of stop codon recognition and the concomitant strict coordination of peptidyl-tRNA hydrolysis (10,11,17). The structure of RF1 bound to a pre-termination complex revealed that the Gln side chain from the GGQ motif may directly coordinate the catalytic water molecule through its carbonyl oxygen, confirming previous molecular dynamics studies (10,18). Methylation of this side chain would stabilize the entire hydrogen bonding network within the PTC and improve RF1/RF2 affinity for the ribosome by increasing hydrophobic interactions (10), rationalizing previous in vitro experiments showing that over-expressed RF2 had a lower affinity to the ribosome than the methylated factor (19).
Methylation of the GGQ motif is conserved in both Saccharomyces cerevisiae and mammalian eRF1 proteins (5,20). This modification is performed by a heterodimeric holoenzyme: Mtq2/Trm112, Pred28α (N6amt1)/mTrm112 and HEMK2α/hTRM112 in yeast, mouse and man, respectively (hereafter referred to as Mtq2-Trm112) (5,20,21). In this complex, the Mtq2 subunit binds the SAM cofactor and catalyses methyl transfer while the Trm112 partner stabilizes and activates Mtq2 (21,22). Trm112 is a small protein composed of two domains: a conserved zinc-binding domain formed by both N- and C-terminal extremities and a central helical domain specific to eukaryotes (22). The Mtq2-Trm112 substrate is the eRF1-eRF3-GTP (or any non-hydrolysable GTP analogue) complex, where eRF3 is the class II release factor of the translational GTPase family, which assists eRF1 in peptide release by inducing a rearrangement of the termination complex upon GTP hydrolysis (22–24). As in prokaryotes, the role of this methylation seems to be associated with the ribosome environment, since methylation should not affect the intrinsic structure of eRF1 (25). Deletion of the MTQ2 gene in S. cerevisiae affects growth (2-fold decrease in growth rate at 30°C) and leads to sensitivity to the antibiotic paromomycin, implying a translation defect related to ribosomal A site function (26) (V.H.H. and S.F., unpublished results). In addition, inactivation of the murine N6amt1 gene leads to early embryonic lethality probably due to cell cycle defects (20). The S. cerevisiae trm112-Δ strains are sicker than mtq2-Δ strains, indicating that Trm112 has additional functions (27) (V.H.H. and S.F., unpublished results). In parallel, inactivation of SMO2, encoding the Trm112 homolog in Arabidopsis thaliana, leads to a defect in progression of cell division and organ growth (28).
Besides Mtq2, Trm112 interacts with and activates two other MTases: Trm9 and Trm11 (22,27,29–32). The Trm9-Trm112 complex catalyses the addition of a methyl group to uridine at wobble position (U34) of some tRNAs (tRNAARG(UCU) and tRNAGLU(UUC)) (33,34). These modified tRNAs are the specific targets of zymocin, a tRNAse toxin secreted by Kluyveromyces lactis (35). Consequently, disruption of TRM9 or TRM112 genes renders strains resistant to that toxin (36). In addition, yeast Trm9 has been implicated in cell death protection by enhancing the translation of DNA damage response key proteins (37). The Trm11-Trm112 complex methylates guanine at position 10 on several yeast tRNAs, forming 2-methylguanosine (30,31).
To gain insight into the role of Trm112 in the activation of these three MTases, we have solved the crystal structure of eukaryotic Mtq2-Trm112 MTase bound to its cofactor SAM and performed in vitro biochemical analysis of mutants as well as in vivo studies. This has allowed us to identify an activation mechanism and to propose a model explaining the need for GTP in the methylation reaction. In addition, in vivo mutational analysis supports the idea that yeast Trm9 interacts with Trm112 in a similar way as Mtq2, rationalizing the role of Trm112 as an activating platform for several MTases.
MATERIALS AND METHODS
Cloning, mutagenesis, expression and purification of proteins
Genes encoding Encephalitozoon cuniculi (Ec)-Mtq2 (UniProtKB entry: Q8SRR4), eRF1 and eRF3 were cloned from Ec-GB-M1 genomic DNA (generous gift from Prof. Vivares, Université Blaise Pascal, Clermont-Ferrand, France) into pET9 plasmids with a C-terminal hexahistidine tag. In parallel, a DNA sequence was designed to encode Ec- Trm112 (UniProtKB entry: Q8SUP0) and Mtq2 with a C-terminus His-tag. This fragment was obtained by de novo gene synthesis (GenScript Corporation, Piscataway, NJ, USA) and was further subcloned into pET21-a between NdeI and XhoI sites. Expression of the His-tagged Mtq2-Trm112 complex was done in E. coli BL21 (DE3) Gold strain (Novagen). The 800 ml of culture in 2xYT medium (BIO101 Inc.) containing ampicillin at 50 µg/ml were incubated at 37°C until an OD600 of about 0.6–0.8 and induced with IPTG 0.5 mM final at 28°C for 4 h. The cells were harvested by centrifugation, resuspended in 40 ml of 20 mM Tris–HCl, pH 7.5, 200 mM NaCl, 5 mM β-mercaptoethanol (buffer A) and stored at −20°C. Cell lysis was achieved by sonication. His-tagged complex was purified on a nickel-nitrilotriacetic acid column (Qiagen Inc.) followed by gel filtration on a SuperdexTM 75 (16/60) column (GE Healthcare) equilibrated in buffer A.
Plasmids used for over-expression of yeast proteins were mutagenized following Quick change mutagenesis protocol (Stratagen) and sequenced. Expression and purification of S. cerevisiae and human proteins were done as described previously (21,22).
Yeast strain construction
Genomic cassette insertion through homologous recombination was used for C-terminus epitope-tagging in yeast according to published protocol (38). Point mutations in Trm9-13Myc encoding gene sequence were also introduced by cassette insertion through homologous recombination (39). Detailed experimental procedure is described in Supplementary Data.
Co-immunoprecipitation and western blot
Soluble protein extracts were prepared by the glass bead method starting from 108 to 109 mid-log growing cells. Cells were washed once in water and pellets were frozen in liquid nitrogen and then stored at −20°C. Cells were thawed and resuspended in ice-cold breaking buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 20 mM MgCl2, 5 mM EDTA, 1% NP-40) supplemented with Complete EDTA-free anti-protease cocktail (Roche Diagnostics). After addition of acid-washed glass beads, cells were lysed by 5 pulses of vortexing for 30 s separated by cooling intervals on ice. Soluble fractions were collected after centrifugation for 20 min at 4°C at 13 000 rpm. Based on Bradford assay, 500 µg of proteins were incubated with mouse monoclonal 9E10 anti-myc antibody (Santa Cruz Biotechnology) for 1 h on ice. Immune complexes were then incubated for 30 min at 4°C on a rotating wheel after addition of protein G-agarose (Sigma). Agarose beads were washed four times in breaking buffer. The final pellet was resuspended in cracking buffer (40 mM Tris–HCl, pH 6.8, 8 M Urea, 5% SDS, 0.1 mM EDTA, 1% β-mercaptoethanol, 0.04% Bromophenol Blue) (40) and boiled for protein release from beads. Proteins from starting soluble fraction and from final supernatant were resolved on a 15% polyacrylamide gel and transferred onto Protran nitrocellulose membrane (Whatman). Probing was performed using either mouse 9E10 anti-myc or 12CA5 anti-HA (Santa Cruz Biotechnology) as primary antibody (1/500) and sheep anti-mouse HRP-conjugated IgG as secondary antibody (1/3000, GE HealthCare). Immunoblots were developed using ECL (GE HealthCare).
Zymocin killer assay
This assay was performed as previously described (41). Procedure is described in Supplementary Data.
Crystallization and structure solution
Thin needle crystals were grown at 19°C from a 0.1:0.1 µl mixture of 7 mg/ml complex solution with a crystallization solution containing 20% poly ethylene glycol (PEG) 4000, 10% isopropanol, 100 mM HEPES (pH 7.5). For data collection, the crystals were transferred into a cryoprotectant crystallization solution with progressively higher ethylene glycol concentrations up to 30% v/v. The datasets were collected at the Zn-edge on beam line Proxima-1 (SOLEIL, St-Aubin, France). The structure was determined by the SAD method (Single wavelength Anomalous Dispersion) using the anomalous signal from the Zn element (see Supplementary Data for details).
The final model contains all the Mtq2 residues (from Met1 to Ser164) as well as residues 1–32 and 36–125 from Trm112, which are well defined in electron density. In addition, one zinc atom, one SAM molecule and 136 water molecules have been modelled. Statistics for data collection and refinement are summarized in Supplementary Table S1. The atomic coordinates and structure factors have been deposited into the Brookhaven Protein Data Bank under the accession number 3Q87.
In vitro methylation assay
Methylation assays were done as described previously by incubating eRF1, eRF3 and GTP with Mtq2-Trm112 complex in presence of [3H] SAM (22).
SAM-binding to human Mtq2 (Hs-Mtq2)
An equimolar amount of Hs-Trm112 was added to partially purified Hs-Mtq2 (corresponding approximately to 100 pmol) and 0.5 µl of [3H] SAM (78 Ci/mmol, Perkin Elmer) in 50 µl final volume methylation buffer for 15 min at 30°C. Reaction was stopped by filtration through nitrocellulose filters (0.45 µm HAWP02500 Millipore) followed by washing with methylation buffer. Radioactivity on filter then represents an amount of SAM bound to Hs-Mtq2. Controls with Hs-Trm112 alone showed very low background fixation. We checked by western blot that Hs-Trm112 was not increasing binding of Hs-Mtq2 itself to the filter, increasing artifactually SAM-binding.
RESULTS
Crystal structure
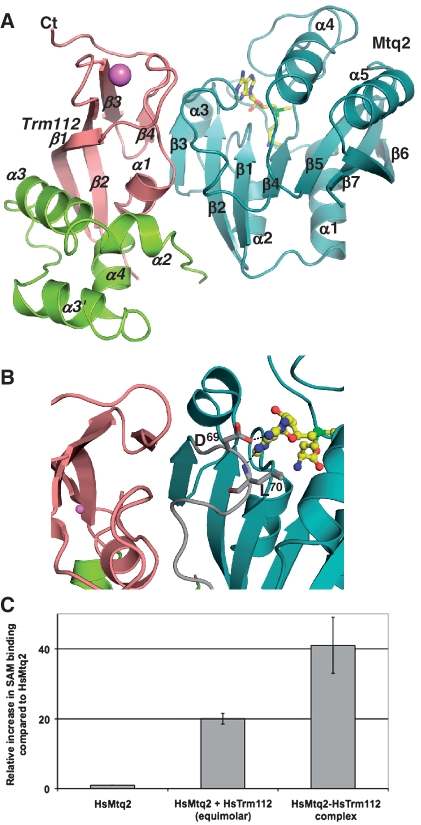
To understand the molecular basis responsible for the interaction between Mtq2 and Trm112, we have crystallized the Mtq2-Trm112 complex from the small parasite E. cuniculi (Ec-Mtq2-Trm112, see ‘Cloning, mutagenesis, expression and purification of proteins’ for more details). We took advantage of the presence of a zinc atom bound to Trm112 as observed for the S. cerevisiae protein (22) to solve the structure of this complex by the SAD method using the zinc anomalous signal. The final model has been refined to 2.1 Å resolution and contains one Mtq2-Trm112 heterodimer in the crystal asymmetric unit (Figure 1A, Supplementary Table S1 and Figure S1 for electron density map). Although we did not pre-incubate the Mtq2-Trm112 complex with SAM for the crystallization experiments, an unambiguous electron density corresponding to a SAM molecule is present in its expected binding site, indicating that this ligand was co-purified with the complex. SAM is bound at the C-terminal face of the Mtq2 central β-sheet and interacts exclusively with residues from Mtq2.
Figure 1.
Mtq2 activation by Trm112. (A) Ribbon representation of the E. cuniculi Mtq2 (blue)-Trm112 complex. The Trm112 zinc-binding and central domains are shown in pink and green, respectively. The purple sphere depicts the zinc atom bound to Trm112. The SAM cofactor bound to Mtq2 is shown as yellow sticks. Secondary structure elements are indicated. Labels in italics correspond to Trm112. (B) Activation loop. Same colour code as panel (A). The loop connecting strands β3 to β4 from Mtq2 is shown in grey. Black dashed lines depict hydrogen bonds. The SAM cofactor bound to Mtq2 is shown as yellow ball and sticks. (C) Effect of HsTrm112 on SAM-binding by HsMtq2. The lower SAM-binding activity observed for the complex formed by the two proteins purified separately is probably due to the instability of the HsMtq2 protein. Hence, contrary to the co-expressed and copurified HsMtq2-Trm112 complex, which is very stable, a significant fraction of HsMtq2 should not be functional in this assay.
Mtq2 adopts the typical class I SAM-dependent MTase fold composed of a central seven-stranded β-sheet (strand order: β3↑β2↑β1↑β4↑β5↑β7↓β6↑) surrounded by three α-helices (α1–α3) on one side and two (α4 and α5) on the other (42). A search for proteins with similar three-dimensional structure [program DALI (43)] identified the C-terminal domain MTase from the PrmC/HemK as the best hit (Z-score = 18.5; RMSD of 2.2 Å over 159 Cα atoms, 19% sequence identity) (14,44). Interestingly, this enzyme performs the same chemical reaction as Mtq2, since it modifies the Gln side chain of the GGQ motif from bacterial release factors RF1 and RF2. The bacterial PrmC MTases are formed by an N-terminal helical domain and a C-terminal MTase domain connected by a short β-hairpin. As anticipated by sequence analysis (5), only the MTase domain is present in Mtq2. In proteins from both kingdoms, these domains are very similar and bind the SAM/S-adenosyl-l-homocysteine (SAH) cofactor in the same way.
The Ec-Trm112 protein is composed of two domains: a zinc-binding domain formed by both N- and C-terminal extremities and a central domain. Its overall structure is very similar to yeast Trm112 [Z-score = 9.3 and global RMSD of 2.9 Å over 102 Cα atoms, 18% sequence identity; Supplementary Figure S2A, (22)]. The zinc-binding domain consists of an α-helix packed onto a four-stranded antiparallel β-sheet (RMSD with the equivalent domain from yeast Trm112 of 1.4 Å over 57 Cα atoms, 30% sequence identity). Similarly, to yeast Trm112, four cysteine residues coordinate the zinc atom. The central domain is a four-helix bundle in the E. cuniculi protein but it is less similar to yeast Trm112 (three-helix bundle) than the zinc-binding domain (RMSD of 3.4 Å over 42 Cα atoms, 5% sequence identity). In this domain, only helices α3 and α4 from Ec-Trm112 match with the equivalent helices from yeast Trm112. Helix α2 from the yeast protein structurally matches with the elongated stretch preceding helix α2 in Ec-Trm112. Helix α3′ from Ec-Trm112 corresponds to the linker connecting helices α3 and α4 in yeast protein. Another important difference between the E. cuniculi and yeast Trm112 structures resides in the orientation of the last ten C-terminal residues that are relatively well conserved. This region was proposed to be part of the homodimer interface in yeast Trm112 and this has since been confirmed by site-directed mutagenesis (data not shown). In the present Mtq2-Trm112 complex, this region cannot adopt the same conformation and folds back onto Trm112 strand β3 to avoid important steric clashes (Supplementary Figure S2B).
Mtq2-Trm112 interface
In the complex, Mtq2 binds mainly to the zinc-binding domain from Trm112 via a β-zipper interaction between Mtq2 strand β3 and Trm112 strand β4 that are associated in a parallel manner (Figure 1A). As a consequence, their β-sheets form a continuous large eleven-stranded β-sheet. Additional contacts are also made by residues from the loop connecting strands β3 to β4 in Mtq2 with helix α1 and the α1-β1 loop from Trm112 (Supplementary Figures S3A–C, S4 and S5). The complete interface buries a total solvent accessible surface area of 2000 Å2 and contains 12 hydrogen bonds, clustered in two regions (see Supplementary Table S2). H-bonds at the β-zipper interface are mostly realized between main chain atoms from both partners (Supplementary Figure S3A). Additional H-bonds occur between residues from loop connecting β1 to α2 in Trm112 and from loop β3-β4 in Mtq2 (Supplementary Figure S3B). Non-polar atoms clustered at the centre of this contact area provide two-thirds of the interface (Supplementary Figure S3B–C). This is indeed illustrated by the strictly conserved Ile125 (S. cerevisiae numbering) from Trm112 strand β4. Its substitution by an aspartic residue results in complete inactivation without disrupting the Mtq2-Trm112 complex (Table 1). A few contacts also exist between strand β2 as well as the β3–β4 loop in Mtq2 and the loop connecting β1 to α2 from the Ec-Trm112 central domain. Helix α2 in yeast Trm112 matches with the region corresponding to the Ec-Trm112 β1-α2 loop and hence is predicted to be important for Mtq2-Trm112 complex formation (Supplementary Figure S3C). This is supported by experiments showing that the yeast complexes containing yeast Trm112 single point mutants F46D and R53E are less soluble and stable than the wild-type complex and have lost eRF1 MTase activity (Table 1). In addition, superposition of yeast Trm112 onto our structure of the complex suggests that additional contacts could occur between helix α2 from Sc-Trm112 and the N-terminal face of the Mtq2 β-sheet as it faces two loops (connecting α1 to β1 and α2 to β2) that are predicted to be longer in other organisms such as S. cerevisiae than in E. cuniculi. This is supported by the N43R mutant, which has decreased eRF1 MTase activity (Supplementary Figure S3C and Table 1).
Table 1.
Effect of Mtq2 and Trm112 mutants on eRF1 methylation in vitro
| eRF1 MTase activity (%) | ||
|---|---|---|
| Trm112 mutants | ||
| N43R (D36) | 58 ± 6 | Located in Ec-Trm112-Mtq2 interface |
| F46D (I39) | 0 | |
| R53E (T46) | 0 | |
| I125D (I115) | 0 | |
| A106E (V96) | 59 ± 11 | Outside from Ec-Mtq2-Trm112 interface |
| E107K (E97) | 61 ± 3 | |
| I118E (I108) | 51 ± 8 | |
| Y120E (P110) | 74 ± 3 | |
| N123R (G113) | 58 ± 1 | |
| Mtq2 mutants | ||
| E16K (E5) | 15 ± 3 | Active site |
| Y15F (Y4) | 0 | |
| E19K (E8) | 0 | |
| D20N (D9) | 0 | |
| F22A (Y11) | 0 | |
| D26K/E29K (D15/E18) | 0 | |
| N122A (N85) | 0 | |
| R207A (R149) | 6.5 ± 2.1 | |
| R207E (R149) | <2 | |
| E212K (E154) | 65 ± 6 | |
| D77A (D51) | 0 | SAM binding |
| D106A/L107A (D69/L70) | 0 | |
All these Mtq2 mutants were co-expressed with wild-type Sc-Trm112 in E. coli and purified using standard protocols. With the exception of the F22A mutant, none of these mutations affected significantly complex formation, solubility and CD spectra (data not shown). Although its CD spectrum was comparable to that of wild-type complex, the Mtq2 (F22A)-Trm112 mutant complex was less stable than wild-type complex. The eRF1 MTase activity of each complex is expressed as a percentage of activity obtained with wild-type enzyme. Absolute activities were measured at least in triplicate, as the initial velocity of the reaction (pmol of eRF1 methylated by second). The E. cuniculi numbering is indicated in parenthesis.
Trm112 enhances SAM-binding by Mtq2
Trm112 is needed to solubilize and activate Mtq2 in yeast (22). Similarly, human Mtq2 (Hs-Mtq2) protein performs eRF1 methylation only after incubation with human Trm112 (21). In order to understand the mechanism of Mtq2 activation by Trm112, we used the structure of the complex to examine the effect of site-directed mutations. Our strategy is based upon two important observations. First, Trm112 shields from the solvent a large hydrophobic surface from Mtq2 by using a hydrophobic region that is involved in yeast Trm112 homodimer formation, explaining why Trm112 solubilizes Mtq2 (Supplementary Figure S3D–E), (22). Second, the loop connecting strands β3–β4 in Mtq2, which is largely involved in the interaction with Trm112, is also involved in SAM-binding (Figure 1B). This led us to speculate that although it does not interact directly with SAM, Trm112 may stabilize this loop in a conformation allowing Mtq2 to bind more tightly the SAM cofactor. In particular, the Asp69 side chain and Leu70 main chain amide groups from this loop are hydrogen-bonded to the adenine moiety of SAM. We have therefore assessed the importance of these two residues for MTase activity by site directed mutagenesis of the corresponding residues in yeast Mtq2 protein (Asp106/Leu107). Co-expression of this double mutant (D106A/L107A) with Trm112 yielded soluble complex, indicating that these mutations do not prevent complex formation. Similarly to the D77A mutant (Asp51 in E. cuniculi), a mutation known to disrupt SAM-binding by MTases, the D106A/L107A, a double mutant is completely unable to methylate eRF1 (Table 1). We next took advantage of the fact that Hs-Mtq2, in contrast with Sc-Mtq2, can be purified in low amounts following over-expression in E. coli in the absence of Hs-Trm112 (21). Although this protein is well folded according to circular dichroism experiments (data not shown), it is unable to methylate eRF1 on its own (21). This could be due to its intrinsically low SAM-binding activity (Figure 1C). Incubation of this protein with an equimolar amount of Hs-Trm112 resulted in restoration of SAM-binding activity (Figure 1C). It is noteworthy that Hs-Trm112 is more efficient when co-expressed with Hs-Mtq2 than upon incubation of the separately purified proteins (Figure 1C). Hence, at least for human proteins, Trm112 strongly stimulates SAM-binding to Mtq2, a prerequisite for enzymatic activity.
MTase active site
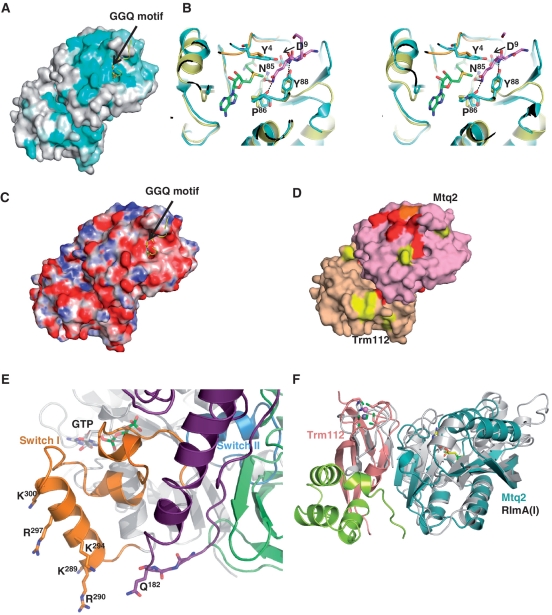
Mapping of sequence conservation at the surface of the Mtq2-Trm112 heterodimer clearly identifies a strongly conserved patch centred on the SAM methyl group as an excellent candidate for the Mtq2 active site (Figure 2A). In addition, the absence of longer loops in orthologous proteins within this putative active site led us to speculate that this structure is an excellent template for the design of mutants from yeast Mtq2 that will affect eRF1 MTase activity.
Figure 2.
Mtq2 active site. (A) Mapping of the sequence conservation at the surface of the Mtq2-Trm112 complex. The RF1 peptide containing the GGQ motif (yellow) has been modelled by superimposing the PrmC-RF1 complex onto Mtq2-Trm112 structure. Colouring is from cyan (highly conserved) to grey (low conservation). (B) Stereo view of the comparison of the Mtq2-Trm112 (blue) and PrmC active sites. The RF1 GGQ motif is shown as pink sticks. The PrmC MTase domain and the linker connecting N-terminal to MTase domains are coloured yellow and orange, respectively. The SAM molecule bound to Mtq2 is shown as green sticks. Hydrogen bonds involved in coordination of the RF1 GGQ motif by PrmC are depicted by dashed lines (14). For clarity, only EcMtq2 residue numbers are indicated. (C) Mapping of the electrostatic potential at the surface of the Mtq2-Trm112 complex. Positively (10 kT/e−) and negatively (−10 kT/e−) charged regions are coloured in blue and red, respectively. Neutral regions are in white. The orientation is the same as in panel (A). (D) Mapping of the Mtq2 and Trm112 residues important for eRF1 methylation in yeast. Mutants affecting partially (>50%), moderately (between 10% and 50%) or completely (<10%) eRF1 methylation are coloured in yellow, orange and red, respectively. The Mtq2 and Trm112 proteins are coloured pink and beige, respectively. Same orientation as panel (A). (E) Model of the eRF1-eRF3-GTP complex. The eRF1 GGQ motif is shown in sticks. For clarity, only the central domain from eRF1 is shown (purple). The GTPase, II and III domains from eRF3 are coloured grey, light green and dark green, respectively. The eRF3 switch regions I and II are coloured orange and blue, respectively. The GTP bound to eRF3 is shown as grey sticks. This model has been generated by superimposing the eRF3 GTPase domain and the eRF1 central domain onto the corresponding domains from the recently solved crystal structure of archaeal Pelota/Dom34-aEF1α-GTP complex (6,24,49). As the switch regions from GTPases are known to adopt the same conformation in the GTP form, we have assumed that the switch regions from aEF1α and eRF3 are similar in the GTP form to model the conformation of this region in the eRF3 GTP bound form. Residues are labelled according to S. cerevisiae numbering. (F) Superimposition of E. coli RlmA(I) (grey) onto Mtq2-Trm112 heterodimer (same colour code as Figure 1A). The residues from RlmA(I) and Trm112 involved in zinc coordination are depicted as sticks. The SAM molecule bound to Mtq2 is shown as yellow sticks. The zinc atoms bound to RlmA(I) and Trm112 are depicted as grey and purple spheres, respectively.
Superposition of the PrmC-RF1 and Mtq2-Trm112 complexes shows that this conserved region corresponds to the bacterial PrmC active site (14). In particular, the Asn-Pro-Pro-Tyr (NPPY) signature found in PrmC but also in DNA N6-adenine and N4-cytosine MTases modifying nitrogens conjugated to planar systems such as an amide group or a nucleotide base (3), matches perfectly the corresponding signature in Mtq2 (Figure 2B). In the complex between PrmC and RF1, this motif forms H-bonds with the Gln side chain of the GGQ motif and positions it ideally for subsequent methylation (14,15). As observed for bacterial PrmC, the Asn122 (Asn85 in Ec-Mtq2) side chain in this NPPY signature is crucial for eRF1 methylation as its substitution by Ala completely abolishes yeast Mtq2 MTase activity (Table 1). We further focused our attention on two strictly conserved residues (Tyr4 and Asp9 in Ec-Mtq2, Tyr15 and Asp20 in yeast) from the N-terminal part of Mtq2 that may participate directly in the coordination of the GGQ motif from eRF1, according to the superposition of the crystal structure of PrmC-RF1 complex onto Mtq2 protein. Both residues are hydrogen-bonded via their side chains and the aromatic ring from Tyr15 packs onto the methyl sulfonio group of the SAM molecule. In order to analyze their role in eRF1 methylation, we have performed conservative substitution of these residues. The Tyr15 side chain was replaced by Phe (Y15F), a substitution that should not much affect SAM-binding. Asp20 was replaced by the isosteric Asn side chain (D20N). Both mutants proved to be completely inactive (Table 1).
In addition, this conserved region displays a negative electrostatic potential that very likely compensates for the positive electrostatic potential surrounding the eRF1 GGQ motif (Figure 2C). We generated several charge inversion mutants of negatively charged residues from this highly conserved putative active site: E16K, E19K, D26K/E29K and E212K (for clarity, yeast numbering is used in this section unless specifically stated). We also tested the following substitutions: F22A, R207A and R207E. Activity measurements show that all the strictly conserved residues located along the solvent exposed face of Mtq2 helix α1 (Glu19, Phe22, Asp26 and Glu29) are crucial for eRF1 methylation, suggesting that this helix is directly involved in substrate recognition (Table 1). This structure-based site-directed mutagenesis approach maps the Mtq2-Trm112 active site to a highly conserved and negatively charged region. This is particularly interesting, since a model of the S. cerevisiae eRF1-eRF3-GTP complex shows that the eRF1 GGQ motif is surrounded by highly conserved and basic residues (see ‘Discussion’ section).
Three additional Mtq2 mutants of solvent-exposed residues affect significantly eRF1 methylation. These are E16K from the loop preceding helix α1 as well as R207A and R207E from strand β6. These residues are located on both sides of helix α1 and hence define a relatively large substrate-binding site. Other mutated positions from Mtq2 (E212K) and Trm112 (A106E, E107K, I118E, Y120E and N123R) affect only partially (50–75%) MTase activity (Table 1). This notably suggests that Trm112 may also participate in substrate binding by this heterodimeric MTase.
The structure of the Mtq2-Trm112 complex can serve as a model for the Trm9-Trm112 complex
Trm112 interacts with and activates three class I SAM-dependent MTases: Mtq2, Trm9 and Trm11. These MTases seem to compete for binding to Trm112, suggesting that they interact with Trm112 in a similar manner (22,27,30–32,36). Hence, our structure of the Mtq2-Trm112 complex may serve as a good model for the Trm9-Trm112 and Trm11-Trm112 complexes and provide a template for functional studies.
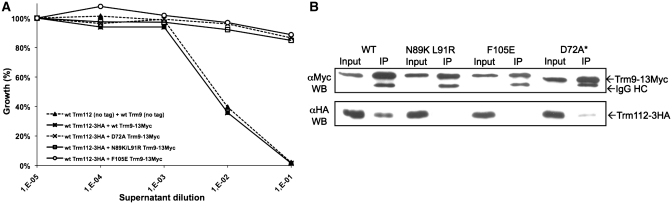
To validate this hypothesis, we have studied the Trm9-Trm112 complex from S. cerevisiae by using K. lactis zymocin toxicity as a tool. This toxin specifically cleaves some tRNAs at the wobble uridine (U34) position. In order to be recognized by the zymocin, these tRNAs need to be fully modified and Trm9 is one of the enzymes involved (45). Hence, this toxin kills wild-type but not S. cerevisiae trm9-Δ or trm112-Δ strains. Using bioinformatics (see Supplementary Data for details on Trm9 model generation), we have generated a model for the S. cerevisiae Trm9 3D-structure and superimposed it onto Mtq2 in the Mtq2-Trm112 complex. We used this model to generate two mutant strains (N89K/L91R and F105E) aimed at disrupting the Trm9-Trm112 interface. As a positive control, we also generated a third mutant strain by substituting the strictly conserved aspartic acid residue proposed to be involved in SAM-binding by Ala (D72A) to inactivate Trm9 as previously done in other MTases (46). We then tested the effect of zymocin addition on cultures of wild-type and mutant yeast strains. All three mutants are resistant to zymocin supporting the hypothesis that these Trm9 mutants are inactive (Figure 3A). Next, the ability of these mutants to interact in vivo with Trm112 was assessed by co-immunoprecipitation. Whereas Trm112 co-immunoprecipitates with wild-type Trm9, no interaction is detected with the N89K/L91R or F105E mutants (Figure 3B). These mutants were expressed at similar levels to wild-type Trm9 protein in vivo, confirming that these mutations have no effect on protein stability. We therefore, conclude that these mutations confer the zymocin resistance phenotype by disrupting the Trm9–Trm112 interaction. However, Trm112 still co-immunoprecipitates with the Trm9 D72A mutant. The zymocin resistance phenotype is here due to the lack of SAM-binding necessary for a normal Trm9 activity. The Trm9–Trm112 interaction is obviously weaker with the Trm9 D72A mutant than with wild-type Trm9, suggesting SAM-binding by Trm9 may influence the strength of interaction between Trm9 and Trm112.
Figure 3.
Saccharomyces cerevisiae Trm9 mutants exhibit induced resistance to zymocin and are affected in in vivo interaction with Trm112. (A) Zymocin killer assay on S. cerevisiae strains. Percentage of growth in the presence of K. lactis AWJ137 supernatant containing zymocin was calculated relative to the control assay performed with supernatant from K. lactis NK40 strain that does not produce zymocin. (B) Effect of Trm9 mutations on Trm9/Trm112 in vivo interaction. Soluble protein extracts (Input: 1/50th of total proteins, i.e. 10 µg) and immunoprecipitates (IP: 1/10th of immunoprecipitated material) were subjected to 15% SDS–PAGE analysis and immunoblotted using anti-Myc (Trm9-13Myc) and anti-HA (Trm112-3HA) as primary antibodies and sheep anti mouse HRP-conjugated IgG as secondary antibody. Similar results were obtained using anti-HA antibodies for protein pull-down (data not shown). The Trm9 D72A mutant protein exhibits a higher electrophoretic mobility due to its reduced copy number of Myc epitopes that is indicated by asterisk (11 instead of 13 for all other Trm9 variants).
Altogether, these experiments strongly support the hypothesis that Trm112 interacts individually with Mtq2, Trm9 and Trm11 and in a similar way. Hence, the structure of the Trm112-Mtq2 complex provides a good model for further mutagenesis studies of the Trm9-Trm112 and Trm11-Trm112 complexes.
DISCUSSION
Post-synthetic modifications are widespread and contribute largely to proteome expansion. Here, we focused our interest on Trm112, a small eukaryotic protein interacting with and activating three SAM-dependent MTases involved in protein synthesis. Two (Trm9 and Trm11) modify tRNAs and the remaining (Mtq2) acts on the class I translation termination factor eRF1 (5,31,34). The crystal structure of the Mtq2-Trm112 complex from E. cuniculi shows that Mtq2 is composed of a single class I SAM-dependent MTase domain also found in PrmC but does not possess the equivalent of the PrmC N-terminal domain involved in RF binding. Superposition of the PrmC-RF1 structure onto the Mtq2-Trm112 complex clearly shows that Trm112 is not a structural stand-in for the PrmC N-terminal domain (Supplementary Figure S6).
Comparison of the active sites of PrmC and Mtq2 shows that the NPPY signature found in MTases modifying nitrogen atoms conjugated to planar systems perfectly superpose and is at the centre of a region highly conserved in eukaryotes. This superposition further shows how the Gln side chain from the eRF1 GGQ motif should be trapped into the Mtq2 active site via a hydrogen bonding network similar to that observed in PrmC-RF1 structure (Figure 2A). Additional structural overlap exists between the loop connecting strand β2 from the short β-hairpin to helix α6 from the MTase domain in PrmC and the N-terminal region preceding helix α1 in Mtq2. In the PrmC-RF1 complex, this loop stacks onto the SAM/SAH molecule and the Gln side chain by contributing a hydrophobic side chain (Leu89 or Phe100 in E. coli or Thermotoga maritima PrmC, respectively). The corresponding residue (Tyr4 or Tyr15 according to E. cuniculi and S. cerevisiae yeast numbering, respectively) is strictly conserved across Mtq2 orthologues. The side chain from Tyr4 lies against the SD atom and methyl donor group from SAM and may form a charge-dipole interaction with the positively charged SD from SAM. In addition, its hydroxyl group is hydrogen-bonded to the strictly conserved Asp9 side chain (Asp20 in S. cerevisiae, Figure 2B and Supplementary Figure S4). Site-directed mutagenesis of these residues in yeast (Y15F or D20N mutants) results in complete loss of eRF1 MTase activity, supporting their crucial role in Mtq2 function (Table 1). A Tyr residue equivalent to Tyr4 from EcMtq2 has also been shown to be important for the activity of the D1 catalytic subunit of the vaccinia virus mRNA capping enzyme (Tyr555) (47).
Next, using site-directed mutagenesis, we have identified highly conserved and negatively charged residues (Glu16, Glu19, Asp26, Asp29 and Glu212) surrounding the NPPY signature from Mtq2 as important for enzymatic activity (Figure 2C and D and Table 1). A model of the S. cerevisiae eRF1-eRF3-GTP complex obtained from the structures of human eRF1-eRF3 (lacking the GTPase domain) (48) and Dom34-Hbs1-GTP complexes (49,50) allows us to propose an explanation for the specificity of Mtq2-Trm112 for the eRF1-eRF3-GTP complex as substrate. First, the GGQ motif from eRF1 central domain is surrounded by several conserved Lys/Arg residues from eRF1 itself. Several of these positively charged residues may be involved in enzyme-substrate complex formation through an interaction with the negatively charged active site of Mtq2. Second, at least one Lys or Arg residue from the eRF1 helix following the GGQ motif (probably Arg189 in S. cerevisiae eRF1 and Arg192 in human eRF1 according to studies conducted on human proteins), (48) should interact with and stabilize the eRF3 switch I region and thereby enhance eRF3 affinity for GTP. This should also stabilize the eRF1 central domain and in particular the GGQ motif and/or optimize the orientation and position of the GGQ motif into Mtq2 active site. Finally, in the eRF1-eRF3-GTP complex, helix α4 from the switch I region (which contains four to five strictly or highly conserved Lys/Arg residues) of the eRF3 GTPase domain is in close proximity from eRF1 GGQ motif and hence, could be involved in Mtq2-Trm112 binding (Figure 2E). As the switch I region from GTPases adopts drastically different conformations depending on the bound guanine nucleotide, this interaction should be specific for the eRF3-GTP form, rationalizing our previous observation that the Mtq2-Trm112 substrate is the eRF1-eRF3 complex bound to GTP but not GDP (5,22). Further studies will be required to determine the mechanism of substrate recognition by Mtq2-Trm112.
Finally, mutations of several solvent exposed Trm112 residues (A106E, E107K, I118E, Y120E and N123R; Table 1) partially affect eRF1 methylation levels without disrupting complex formation and fold (data not shown). Strikingly, the protein displaying the higher structural similarity with the Mtq2-Trm112 complex is the bacterial protein RlmA(I), which is involved in methylation of 23S rRNA at position G745 (51). RlmA(I) is composed of an N-terminal zinc finger domain followed by a C-terminal class I MTase domain (52). Superposition of the RlmA(I) MTase domain onto the Mtq2 protein (RMSD of 2.6 Å over 120 Cα atoms, 7% sequence identity) in our complex reveals that the Trm112 zinc finger domain nicely matches with the RlmA(I) zinc finger domain that has been suggested to interact with the rRNA substrate (Figure 2F) (52). This suggests that Trm112 may assist Mtq2 in substrate recognition. Such structural similarity with RlmA(I) was previously proposed for the Tmr11-Trm112 complex (RMSD between our crystal structure and the Trm11-Trm112 model is 2.35 Å over 191 Cα atoms) (31).
The Mtq2-Trm112 structure also suggests how Trm112 activates Mtq2 and hence its other MTase partners. First, Trm112 masks a hydrophobic region from Mtq2 upon complex formation, explaining the need for co-expression of Trm112 and Mtq2 to solubilize Mtq2 (Supplementary Figure 3D and E) (22). The same mechanism may be shared by Trm9, which can only be recovered as a soluble protein upon co-expression with Trm112 (22,27) (M.G. and V.H.H., unpublished data). Second, Trm112 stimulates SAM-binding by Mtq2 via its interaction with the Mtq2 loop connecting strands β3–β4, which in turn contacts the SAM molecule (Figure 1B and C). Moreover, the Trm9 mutant (D72A) predicted to be deficient in SAM-binding interacts more weakly with Trm112 than wild-type Trm9 (Figure 3B). Altogether, this strongly argues in favour of a synergistic interaction between SAM, Trm112 and Mtq2/Trm9. We project that Trm112 may assist substrate binding by its three partners. Indeed, as mentioned above, we have identified mutations of solvent-exposed Trm112 residues affecting eRF1 methylation by Mtq2 (Table 1). However, the implication of Trm112 in the recognition of substrates as different as tRNA and eRF1/eRF3 will need to be further addressed. The requirement for a protein partner to activate MTases is not unique to Trm112 but was described for the following holoenzymes Trm8-Trm82, Trm61-Trm6, vaccinia virus D1-D12 capping enzyme (46,53–57). The vaccinia virus D12 activator subunit stabilizes the D1 subunit, increases its catalytic activity and its affinity for both its substrate and SAM (57). Regarding the Trm8-Trm82 complex, which catalyses m7G methylation at position 46 of some tRNAs (58), Trm82 is not directly involved in tRNA binding but should rather stabilize Trm8 by modulating the structure of its catalytic site (53,59). In the Trm61-Trm6 complex, Trm6 is required for tRNA binding and hence catalysis of A58 methylation by the Trm61 MTase (54,56).
Several observations support the hypothesis that Trm112 interacts in a similar way with its three MTase partners. First, sequence analysis predicts that Mtq2, Trm9 and Trm11 belong to the same structural family of class I SAM-dependent MTases (3,31,34). Second, over-expression of Mtq2 or Trm11 affects the interaction between Trm9 and Trm112, thereby inhibiting Trm9 activity and consequently conferring resistance to zymocin (36). This is in line with previous results indicating that Trm112 interacts independently with its partners (31). Third, the β-zipper type of interaction between strands β4 from Trm112 and β3 from Mtq2 (Figure 1A and Supplementary Figure 3A) involves hydrogen bonds between main chain atoms and may therefore be well adapted for interaction with proteins with totally different sequences but similar folds. Fourth, mutations of Trm9 residues (N89K/L91R and F105E) corresponding to Mtq2 residues involved in the interface with Trm112 confer resistance to zymocin (i.e. inactivate Trm9) and disrupt interaction with Trm112 (Figure 3A and B). Altogether, these observations strongly argue in favour of a similar interaction mode between Trm112 and its three MTase partners. It is noteworthy that the Trm9–Trm112 interaction is conserved in human, since the tRNA MTase domain from human Trm9 also interacts with human Trm112 (29,32). Similarly, Mtq2 and Trm112 proteins are conserved in eukaryotes and archaea and their interaction has been shown in yeast and mammals. Their deletion has a strong impact on growth and development but it is not known if this is a direct consequence of the absence of eRF1 methylation, leading to a termination defect during protein synthesis. Indeed, the absence of methylation does not influence significantly in vivo readthrough in yeast and could even have an opposite effect (26) (VHH, SF unpublished results). In addition, it has been shown that increased stop codon readthrough in eRF1 mutants or [PSI+] and yeast variants does not lead to a reduction in growth rate (60–62). In yeast, TRM112 deletion is more detrimental than the combined deletions of known partners (Mtq2, Trm9, Trm11), suggesting another function for Trm112. Preliminary results suggest that an interaction between Trm112 and Sfh1, an essential component of the RSC complex involved in transcription and necessary for proper mitosis, might be biologically relevant (27). Clearly, more studies aimed at deciphering the role of Trm112 are needed and our crystal structure of the Mtq2-Trm112 complex provides an excellent template for designing future experiments towards this goal.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Agence Nationale pour la Recherche (grant ANR-06-BLAN-0075-02 and ANR-07-JCJC-0105); the Centre National pour la Recherche Scientifique; the Human Frontier Science Program organism (grant RGP0018/2009-C); the EU ‘3D-Repertoire’ program (LSHG-CT-2005-512028); pre-doctoral grant from the Université Paris-Sud 11 (to J.H.). Funding for open access charge: Human Frontier Science Program organism (grant RGP0018/2009-C).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to J. Cicolari for technical assistance. We thank Tony Johnson (NIMR, Mill Hill, London) for the generous gift of S. cerevisiae CG378 strain and plasmids pFA6a-3HA-TRP1 and pFA6a-13Myc-kanMX6, as well as Dr K. Breunig and Dr R. Schaffrath for sharing with us the K. lactis AWJ137 and NK40 strains, respectively. We acknowledge SOLEIL for provision of synchrotron radiation facilities and we would like to thank Dr Andrew Thompson and Pierre Legrand for assistance with beam line Proxima-1. We are indebted to Dr B. Lapeyre and J.M. Bujnicki for sharing with us the coordinates of their Trm11-Trm112 model.
REFERENCES
- 1.Pang CN, Gasteiger E, Wilkins MR. Identification of arginine- and lysine-methylation in the proteome of Saccharomyces cerevisiae and its functional implications. BMC Genomics. 2010;11:92. doi: 10.1186/1471-2164-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polevoda B, Sherman F. Methylation of proteins involved in translation. Mol. Microbiol. 2007;65:590–606. doi: 10.1111/j.1365-2958.2007.05831.x. [DOI] [PubMed] [Google Scholar]
- 3.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dincbas-Renqvist V, Engstrom A, Mora L, Heurgue-Hamard V, Buckingham R, Ehrenberg M. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 2000;19:6900–6907. doi: 10.1093/emboj/19.24.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heurgue-Hamard V, Champ S, Mora L, Merkulova-Rainon T, Kisselev LL, Buckingham RH. The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J. Biol. Chem. 2005;280:2439–2445. doi: 10.1074/jbc.M407252200. [DOI] [PubMed] [Google Scholar]
- 6.Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. The crystal structure of human eukaryotic release factor eRF1–mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 7.Vestergaard B, Van LB, Andersen GR, Nyborg J, Buckingham RH, Kjeldgaard M. Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol. Cell. 2001;8:1375–1382. doi: 10.1016/s1097-2765(01)00415-4. [DOI] [PubMed] [Google Scholar]
- 8.Kisselev LL, Buckingham RH. Translational termination comes of age. Trends Biochem. Sci. 2000;25:561–566. doi: 10.1016/s0968-0004(00)01669-8. [DOI] [PubMed] [Google Scholar]
- 9.Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korostelev A, Zhu J, Asahara H, Noller HF. Recognition of the amber UAG stop codon by release factor RF1. EMBO J. 2010;29:2577–2585. doi: 10.1038/emboj.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loh PG, Song H. Structural and mechanistic insights into translation termination. Curr. Opin. Struct. Biol. 2010;20:98–103. doi: 10.1016/j.sbi.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Heurgué-Hamard V, Champ S, Engstöm Å, Ehrenberg M, Buckingham RH. The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors. EMBO J. 2002;21:769–778. doi: 10.1093/emboj/21.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakahigashi K, Kubo N, Narita S, Shimaoka T, Goto S, Oshima T, Mori H, Maeda M, Wada C, Inokuchi H. HemK, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination. Proc. Natl Acad. Sci. USA. 2002;99:1473–1478. doi: 10.1073/pnas.032488499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graille M, Heurgue-Hamard V, Champ S, Mora L, Scrima N, Ulryck N, van Tilbeurgh H, Buckingham RH. Molecular basis for bacterial class I release factor methylation by PrmC. Mol. Cell. 2005;20:917–927. doi: 10.1016/j.molcel.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 15.Schubert HL, Phillips JD, Hill CP. Structures along the catalytic pathway of PrmC/HemK, an N5-glutamine AdoMet-dependent methyltransferase. Biochemistry. 2003;42:5592–5599. doi: 10.1021/bi034026p. [DOI] [PubMed] [Google Scholar]
- 16.Mora L, Heurgue-Hamard V, de Zamaroczy M, Kervestin S, Buckingham RH. Methylation of bacterial release factors RF1 and RF2 is required for normal translation termination in vivo. J. Biol. Chem. 2007;282:35638–35645. doi: 10.1074/jbc.M706076200. [DOI] [PubMed] [Google Scholar]
- 17.Jin H, Kelley AC, Loakes D, Ramakrishnan V. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc. Natl Acad. Sci. USA. 2010;107:8593–8598. doi: 10.1073/pnas.1003995107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trobro S, Aqvist J. A model for how ribosomal release factors induce peptidyl-tRNA cleavage in termination of protein synthesis. Mol. Cell. 2007;27:758–766. doi: 10.1016/j.molcel.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 19.Pavlov MY, Freistroffer DV, Dincbas V, MacDougall J, Buckingham RH, Ehrenberg M. A direct estimation of the context effect on the efficiency of termination. J. Mol. Biol. 1998;284:579–590. doi: 10.1006/jmbi.1998.2220. [DOI] [PubMed] [Google Scholar]
- 20.Liu P, Nie S, Li B, Yang ZQ, Xu ZM, Fei J, Lin C, Zeng R, Xu GL. Deficiency in a glutamine-specific methyltransferase for the release factor causes mouse embryonic lethality. Mol. Cell. Biol. 2010;30:4245–4253. doi: 10.1128/MCB.00218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figaro S, Scrima N, Buckingham RH, Heurgue-Hamard V. HemK2 protein, encoded on human chromosome 21, methylates translation termination factor eRF1. FEBS Lett. 2008;582:2352–2356. doi: 10.1016/j.febslet.2008.05.045. [DOI] [PubMed] [Google Scholar]
- 22.Heurgue-Hamard V, Graille M, Scrima N, Ulryck N, Champ S, van Tilbeurgh H, Buckingham RH. The zinc finger protein Ynr046w is plurifunctional and a component of the eRF1 methyltransferase in yeast. J. Biol. Chem. 2006;281:36140–36148. doi: 10.1074/jbc.M608571200. [DOI] [PubMed] [Google Scholar]
- 23.Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 24.Kong C, Ito K, Walsh MA, Wada M, Liu Y, Kumar S, Barford D, Nakamura Y, Song H. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol. Cell. 2004;14:233–245. doi: 10.1016/s1097-2765(04)00206-0. [DOI] [PubMed] [Google Scholar]
- 25.Ander M, Aqvist J. Does glutamine methylation affect the intrinsic conformation of the universally conserved GGQ motif in ribosomal release factors? Biochemistry. 2009;48:3483–3489. doi: 10.1021/bi900117r. [DOI] [PubMed] [Google Scholar]
- 26.Polevoda B, Span L, Sherman F. The yeast translation release factors Mrf1p and Sup45p (eRF1) are methylated, respectively, by the methyltransferases Mtq1p and Mtq2p. J. Biol. Chem. 2006;281:2562–2571. doi: 10.1074/jbc.M507651200. [DOI] [PubMed] [Google Scholar]
- 27.Mazauric MH, Dirick L, Purushothaman SK, Bjork GR, Lapeyre B. Trm112p is a 15-kDa zinc finger protein essential for the activity of two tRNA and one protein methyltransferases in yeast. J. Biol. Chem. 2010;285:18505–18515. doi: 10.1074/jbc.M110.113100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Z, Qin Z, Wang M, Xu C, Feng G, Liu J, Meng Z, Hu Y. The Arabidopsis SMO2, a homologue of yeast TRM112, modulates progression of cell division during organ growth. Plant J. 2010;61:600–610. doi: 10.1111/j.1365-313X.2009.04085.x. [DOI] [PubMed] [Google Scholar]
- 29.Fu D, Brophy JA, Chan CT, Atmore KA, Begley U, Paules RS, Dedon PC, Begley TJ, Samson LD. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell. Biol. 2010;30:2449–2459. doi: 10.1128/MCB.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada K, Muneyoshi Y, Endo Y, Hori H. Production of yeast (m2G10) methyltransferase (Trm11 and Trm112 complex) in a wheat germ cell-free translation system. Nucleic Acids Symp. Ser. 2009;53:303–304. doi: 10.1093/nass/nrp152. [DOI] [PubMed] [Google Scholar]
- 31.Purushothaman SK, Bujnicki JM, Grosjean H, Lapeyre B. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol. Cell. Biol. 2005;25:4359–4370. doi: 10.1128/MCB.25.11.4359-4370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Songe-Moller L, van den Born E, Leihne V, Vagbo CB, Kristoffersen T, Krokan HE, Kirpekar F, Falnes PO, Klungland A. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol. Cell. Biol. 2010;30:1814–1827. doi: 10.1128/MCB.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang B, Lu J, Bystrom AS. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA. 2008;14:2183–2194. doi: 10.1261/rna.1184108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalhor HR, Clarke S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol. Cell. Biol. 2003;23:9283–9292. doi: 10.1128/MCB.23.24.9283-9292.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jablonowski D, Zink S, Mehlgarten C, Daum G, Schaffrath R. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator's key role for zymocin-induced cell death in yeast. Mol. Microbiol. 2006;59:677–688. doi: 10.1111/j.1365-2958.2005.04972.x. [DOI] [PubMed] [Google Scholar]
- 36.Studte P, Zink S, Jablonowski D, Bar C, von der Haar T, Tuite MF, Schaffrath R. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol. Microbiol. 2008;69:1266–1277. doi: 10.1111/j.1365-2958.2008.06358.x. [DOI] [PubMed] [Google Scholar]
- 37.Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 39.Toulmay A, Schneiter R. A two-step method for the introduction of single or multiple defined point mutations into the genome of Saccharomyces cerevisiae. Yeast. 2006;23:825–831. doi: 10.1002/yea.1397. [DOI] [PubMed] [Google Scholar]
- 40.Printen JA, Sprague GF., Jr Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolf K, Breunig K, Barth G. Non-conventional Yeasts in Genetics, Biochemistry and Biotechnology: Practical Protocols Chapter 28. Berlin, New York: Springer; 2003. pp. 179–183. [Google Scholar]
- 42.Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 43.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Z, Shipman L, Zhang M, Anton BP, Roberts RJ, Cheng X. Structural characterization and comparative phylogenetic analysis of Escherichia coli HemK, a protein (N5)-glutamine methyltransferase. J. Mol. Biol. 2004;340:695–706. doi: 10.1016/j.jmb.2004.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu J, Huang B, Esberg A, Johansson MJ, Bystrom AS. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leulliot N, Bohnsack MT, Graille M, Tollervey D, Van Tilbeurgh H. The yeast ribosome synthesis factor Emg1 is a novel member of the superfamily of alpha/beta knot fold methyltransferases. Nucleic Acids Res. 2008;36:629–639. doi: 10.1093/nar/gkm1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De la Pena M, Kyrieleis OJ, Cusack S. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. EMBO J. 2007;26:4913–4925. doi: 10.1038/sj.emboj.7601912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng Z, Saito K, Pisarev AV, Wada M, Pisareva VP, Pestova TV, Gajda M, Round A, Kong C, Lim M, et al. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 2009;23:1106–1118. doi: 10.1101/gad.1770109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi K, Kikuno I, Kuroha K, Saito K, Ito K, Ishitani R, Inada T, Nureki O. Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1{alpha} complex. Proc. Natl Acad. Sci. USA. 2010;107:17575–17579. doi: 10.1073/pnas.1009598107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van den Elzen AM, Henri J, Lazar N, Gas ME, Durand D, Lacroute F, Nicaise M, van Tilbeurgh H, Seraphin B, Graille M. Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways. Nat. Struct. Mol. Biol. 2010;17:1446–1452. doi: 10.1038/nsmb.1963. [DOI] [PubMed] [Google Scholar]
- 51.Liu M, Douthwaite S. Resistance to the macrolide antibiotic tylosin is conferred by single methylations at 23S rRNA nucleotides G748 and A2058 acting in synergy. Proc. Natl Acad. Sci. USA. 2002;99:14658–14663. doi: 10.1073/pnas.232580599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Das K, Acton T, Chiang Y, Shih L, Arnold E, Montelione GT. Crystal structure of RlmAI: implications for understanding the 23S rRNA G745/G748-methylation at the macrolide antibiotic-binding site. Proc. Natl Acad. Sci. USA. 2004;101:4041–4046. doi: 10.1073/pnas.0400189101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexandrov A, Grayhack EJ, Phizicky EM. tRNA m7G methyltransferase Trm8p/Trm82p: evidence linking activity to a growth phenotype and implicating Trm82p in maintaining levels of active Trm8p. RNA. 2005;11:821–830. doi: 10.1261/rna.2030705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anderson J, Phan L, Hinnebusch AG. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2000;97:5173–5178. doi: 10.1073/pnas.090102597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mao X, Shuman S. Intrinsic RNA (guanine-7) methyltransferase activity of the vaccinia virus capping enzyme D1 subunit is stimulated by the D12 subunit. Identification of amino acid residues in the D1 protein required for subunit association and methyl group transfer. J. Biol. Chem. 1994;269:24472–24479. [PubMed] [Google Scholar]
- 56.Ozanick SG, Bujnicki JM, Sem DS, Anderson JT. Conserved amino acids in each subunit of the heteroligomeric tRNA m1A58 Mtase from Saccharomyces cerevisiae contribute to tRNA binding. Nucleic Acids Res. 2007;35:6808–6819. doi: 10.1093/nar/gkm574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwer B, Hausmann S, Schneider S, Shuman S. Poxvirus mRNA cap methyltransferase. Bypass of the requirement for the stimulatory subunit by mutations in the catalytic subunit and evidence for intersubunit allostery. J. Biol. Chem. 2006;281:18953–18960. doi: 10.1074/jbc.M602867200. [DOI] [PubMed] [Google Scholar]
- 58.Alexandrov A, Martzen MR, Phizicky EM. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA. 2002;8:1253–1266. doi: 10.1017/s1355838202024019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leulliot N, Chaillet M, Durand D, Ulryck N, Blondeau K, van Tilbeurgh H. Structure of the yeast tRNA m7G methylation complex. Structure. 2008;16:52–61. doi: 10.1016/j.str.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 60.Henri J, Rispal D, Bayart E, van Tilbeurgh H, Seraphin B, Graille M. Structural and functional insights into Saccharomyces cerevisiae Tpa1, a putative prolylhydroxylase influencing translation termination and transcription. J. Biol. Chem. 2010;285:30767–30778. doi: 10.1074/jbc.M110.106864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Merritt GH, Naemi WR, Mugnier P, Webb HM, Tuite MF, von der Haar T. Decoding accuracy in eRF1 mutants and its correlation with pleiotropic quantitative traits in yeast. Nucleic Acids Res. 2010;38:5479–5492. doi: 10.1093/nar/gkq338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson MA, Meaux S, Parker R, van Hoof A. Genetic interactions between [PSI+] and nonstop mRNA decay affect phenotypic variation. Proc. Natl Acad. Sci. USA. 2005;102:10244–10249. doi: 10.1073/pnas.0504557102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.