Abstract
Homozygosity for mutations in ABC transporter A1 (ABCA1) causes Tangier disease, a rare HDL-deficiency syndrome. Whether heterozygosity for genetic variation in ABCA1 also contributes to HDL cholesterol (HDL-C) levels in the general population is presently unclear. We determined whether mutations or single-nucleotide polymorphisms (SNPs) in ABCA1 were overrepresented in individuals with the lowest 1% (n = 95) or highest 1% (n = 95) HDL-C levels in the general population by screening the core promoter and coding region of ABCA1. For all nonsynonymous SNPs identified, we determined the effect of genotype on lipid traits in 9,259 individuals from the general population. Heterozygosity for ABCA1 mutations was identified in 10% of individuals with low HDL-C only. Three of 6 nonsynonymous SNPs (V771M, V825I, and R1587K) were associated with increases or decreases in HDL-C in women in the general population and some with consistent trends in men, determined as isolated single-site effects varying only at the relevant SNP. Finally, these results were consistent over time. In conclusion, we show that at least 10% of individuals with low HDL-C in the general population are heterozygous for mutations in ABCA1 and that both mutations and SNPs in ABCA1 contribute to HDL-C levels in the general population.
Introduction
Tangier disease is a rare recessive disorder characterized by almost complete absence of HDL particles in plasma, accumulation of cholesterol esters in macrophage-rich tissues, and increased susceptibility to atherosclerosis (1). The molecular basis for Tangier disease has recently been established as homozygosity for defects in ABC transporter A1 (ABCA1) (2–4). The recognition of ABCA1 mutations as the molecular basis for Tangier disease and the milder heterozygous form, familial hypoalphalipoproteinemia, has contributed substantially to the understanding of the normal function of ABCA1 as a key transporter of cellular cholesterol across cell membranes to acceptor molecules in plasma such as apoAI. Thus, ABCA1 influences the initial steps in HDL formation and in reverse cholesterol transport, potentially important for the development of atherosclerosis (5–7).
Twin and family studies suggest that about 50% of the variation in HDL cholesterol (HDL-C) levels is genetically determined (8, 9). Although defects in several genes such as APOAI, APOE, lipoprotein lipase, hepatic lipase, lechitin cholesterol acyltransferase, and cholesteryl ester transfer protein all are known to influence HDL-C levels (10), these defects account for only a small proportion of genetic variation in HDL-C. Because ABCA1 is crucial in the initial step of HDL formation and in reverse cholesterol transport, and because ABCA1 mutations in the homozygous form cause a rare HDL-deficiency syndrome, heterozygosity for mutations (rare allele frequency ≤ 1%) and single-nucleotide polymorphisms (SNPs; rare allele frequency > 1%) in ABCA1 may also affect plasma HDL-C levels in the general population.
We tested the following hypotheses: (a) heterozygosity for mutations in ABCA1 is overrepresented in individuals with low HDL-C; (b) frequencies of SNPs in ABCA1 tend to differ between individuals with low and high HDL-C levels; (c) SNPs in ABCA1 affect HDL-C levels in the general population. To increase the likelihood of identifying genetic variation with significant effects on HDL-C levels, we used a systematic approach. We screened the core promoter and all 50 exons of ABCA1 (Ν17 kb) in individuals with the lowest 1% (n = 95) and highest 1% (n = 95) HDL-C levels for age and sex from an ethnically homogeneous general population sample, The Copenhagen City Heart Study (n = 9,259). All nonsynonymous SNPs thus identified were genotyped in the entire general population sample, the effect on lipid and lipoprotein levels was determined as overall effects (regardless of variation at the other sites) and as isolated single site effects (genotypes differing only at the relevant SNP), and finally the results were verified at 2 examinations 10 years apart.
Results
Characteristics of participants.
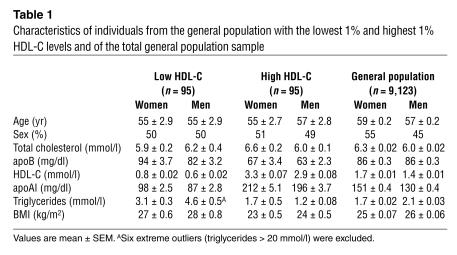
Characteristics of individuals in the low HDL-C group, the high HDL-C group, and the entire general population sample are shown in Table 1. Individuals in the low HDL-C group had lower apoAI levels, higher triglyceride levels, and were more obese than individuals in the high HDL-C group or in the general population (Table 1).
Table 1.
Characteristics of individuals from the general population with the lowest 1% and highest 1% HDL-C levels and of the total general population sample
Allele frequencies.
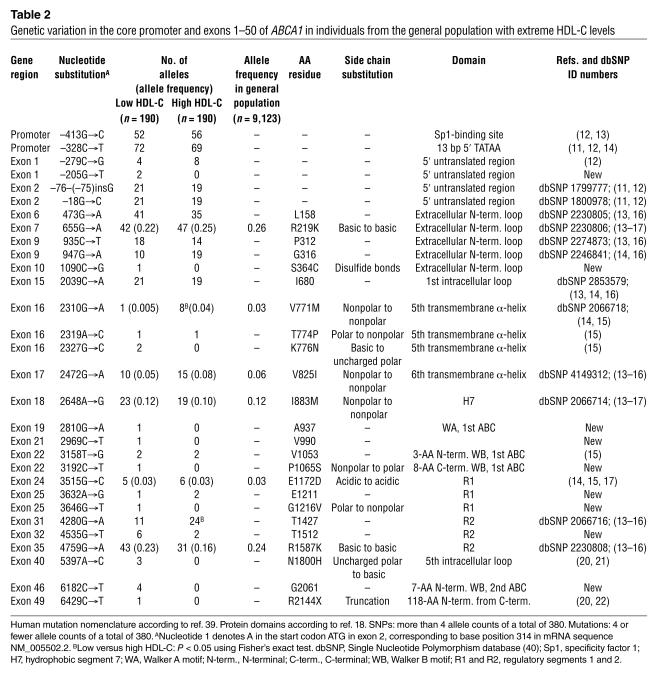
A total of 51 genetic variants were identified. Allele frequencies of the 30 genetic variants identified in regulatory and transcribed parts of ABCA1 ranged from 1 of 380 alleles (0.003) to 141 of 380 alleles (0.37) in the low and high HDL-C groups (Table 2). Seventeen SNPs (allele frequencies > 1%) were identified in both HDL-C groups, and the most common of these have been reported previously. The frequencies of 3 of these SNPs differed significantly (V771M, P = 0.04; T1427, P = 0.02) or were borderline (R1587K, P = 0.12) between individuals with the lowest 1% and highest 1% HDL-C levels. Of the 13 mutations (allele frequencies ≤ 1%), 9 were identified exclusively in individuals with the lowest 1% HDL-C levels (Table 2). Allele frequencies of the 22 intron variants ranged from 1/190 alleles (0.5%) to 40/190 alleles (21%) (see Supplemental Table 2; supplemental material available at http://www.jci.org/cgi/content/full/114/9/1343/DC1).
Table 2.
Genetic variation in the core promoter and exons 1–50 of ABCA1 in individuals from the general population with extreme HDL-C levels
Allele frequencies of the 6 nonsynonymous SNPs (R219K, V771M, V825I, I883M, E1172D, R1587K) that were genotyped in the total general population sample ranged from 0.03 (V771M and E1172D) to 0.26 (R219K) (Table 2).
Genetic variation in the core promoter and 5 ′ untranslated region.
Five SNPs and 1 mutation were identified in the core promoter and 5′ untranslated region (5′ UTR) of exons 1 and 2 (Table 2). The –205GØT mutation, which has not been reported previously, was identified in 2 individuals with the lowest 1% HDL-C levels. By contrast, the 5 SNPs have already been reported (11–14); none of these differed in frequency between individuals with low and high HDL-C.
SNPs in coding regions.
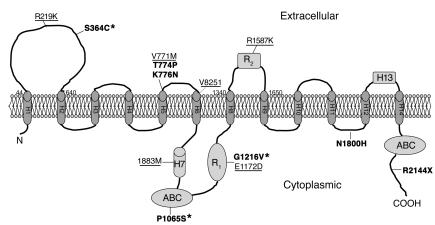
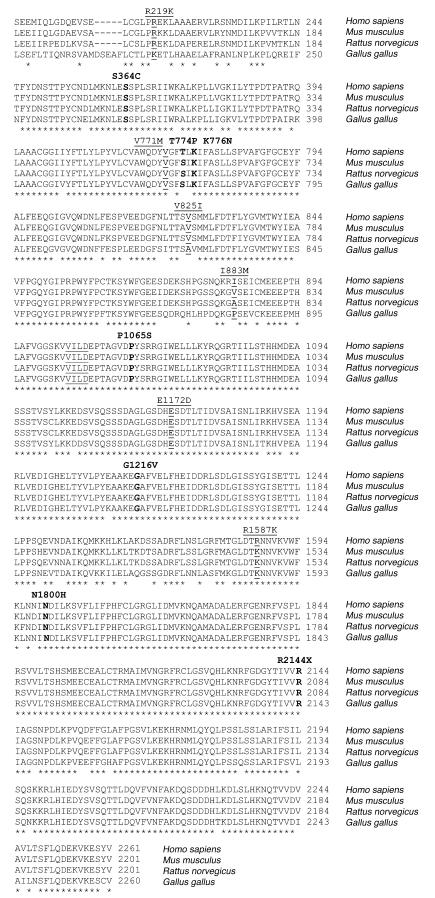
Twelve SNPs were identified in coding regions of the gene, 6 of which resulted in amino acid substitutions (Table 2). R219K, V771M, V825I, I883M, E1172D, and R1587K all substitute similar amino acids. R219K and R1587K are located in the 2 major extracellular loops of the ABCA1 protein, important for the interaction with apoAI and for cholesterol efflux, whereas V771M, V825I, I883M, and E1172D are located in the middle part of the protein corresponding to the fifth and sixth transmembrane α-helix, the seventh hydrophobic segment (H7), and the first regulatory segment (R1), respectively (Table 2, Figure 1). V771M was more frequent in the high HDL-C group (P = 0.04), whereas R1587K tended to be overrepresented in the low HDL-C group (P = 0.12). The amino acid residues affected by these 6 nonsynonymous SNPs are situated in highly conserved (V771, V825, E1172, R1587) or less-conserved (R219, I883) areas of ABCA1 and are either completely conserved between species (V771, E1172) or vary between 2 (R219, V825, R1587) or more (I883) similar amino acids (Figure 2). All 6 nonsynonymous SNPs are common and have been reported previously (13–17).
Figure 1.
Topological model of ABCA1 according to Fitzgerald et al. (18). The amino acid substitutions identified in the present study are superimposed (SNPs are underlined, mutations are in bold). Location of the variants is deduced from ref. 18. ABC transporters are composed of 4 parts: 2 membrane-integral domains (H1–H6 and H8–H12 + H14), each of which spans the membrane 6 times, and 2 ATP-hydrolyzing domains (ABCs), which contain the highly conserved Walker A (GXXGXGKS/T) and B motifs (hhhhD; h, hydrophobic amino acid) connected by an ABC family–specific signature motif (LSGGQQ/R/KQR) (38). H1–H13, hydrophobic segment 1 to 13; R1 and R2, regulatory segments 1 and 2. Asterisks indicate new mutations identified in the present study.
Figure 2.
Alignment of ABCA1 between species. Human ABCA1 (NP_005493.2) and the orthologous murine (NP_038482.1), rat (NP_835196.1), and chicken (AAL56247.1) protein sequences were aligned with the ClustalW program (http://www.ebi.ac.uk/clustalw/). Selected parts of the protein with the identified amino acid substitutions are marked in underscored (SNPs) and bold (mutations), respectively. Underscored amino acids (VVILD) at residues 1053–1057 mark the Walker B domain. At residue 1587, K (A allele) is reported to be the WT in humans (GenBank protein accession number NP_005493.2). In this study, R (G allele) is the most common amino acid, and similar results have been reported by others (29). Asterisks indicate identical amino acid residues in all sequences aligned.
The remaining 6 SNPs in the coding region of ABCA1 did not substitute an amino acid. T1427 was significantly more frequent in the high HDL-C group (P = 0.02), most likely due to negative linkage disequilibrium (LD) with R1587K (see Supplemental Table 3). The least frequent of these SNPs, T1512, is new, while L158, P312, G316, I680, and T1427 have been reported by others (13–16).
Mutations in coding regions.
Twelve mutations were identified in coding regions of the gene, 6 of which introduced amino acid substitutions and 1 a premature stop-codon (Table 2): S364C introduces a cysteine that allows the formation of disulfide bonds; T774P, P1065S, and G1216V introduce a shift between polar and nonpolar side chains; K776N and N1800H introduce a shift between basic and uncharged polar side chains; and R2144X results in a truncated protein lacking the C terminal 118 amino acids (Table 2). S364C is situated in the first extracellular loop known to be important for the interaction with apoAI and thus for cholesterol efflux (18, 19) (Table 2 and Figure 1). P1065S is located 8 amino acids downstream of the Walker B motif of the first ABC, and G1216V is in the R1 segment of the large central regulatory region rich in polar and charged residues. These 3 mutations have not been described previously. T774P, K776N, N1800H, and R2144X have been reported by others (15, 20–22); T774P and K776N are located in the fifth transmembrane α-helix; N1800H in the fifth intracellular loop; and R2144X in the intracellular C terminus (Table 2, Figure 1).
The amino acid residues affected by these 7 mutations are situated in highly conserved areas of the ABCA1 protein and are themselves completely conserved between species (human, mouse, rat, chicken), with the exception of T774, which varies between similar amino acids (TØS) (Figure 2). Six of these amino acids (again except T774) are also relatively conserved between most human ABCAs, either due to the presence of identical residues (S364, P1065) or the presence of hydrophilic amino acids (K776, G1216, N1800) (Figure 3). The high degree of conservation suggests that these amino acid residues are important for normal function of the ABCA1 protein.
Figure 3.
Alignment of human ABCAs. ABCAs are all full transporters and are involved in sterol transport, either tissue specific or more generalized. The ABCA11 gene sequence/protein sequence is not available in Homo sapiens. ABCA1 to ABCA10 and ABCA12 to ABCA13 Homo sapiens protein sequences, NP_005493.2, NP_001597.1, NP_001080.1, NP_000341.1, NP_061142.2, NP_525023.2, NP_061985.1, NP_009099.1, NP_525022.2, NP_525021.2, NP_775099.2, NP_689914.2, were aligned with the ClustalW program (http://ebi.ac.uk/clustalw/). Selected parts of the proteins with the identified amino acid substitutions are marked in bold (mutations) and underscored (SNPs), respectively. Asterisks indicate identical amino acid residues in all sequences aligned.
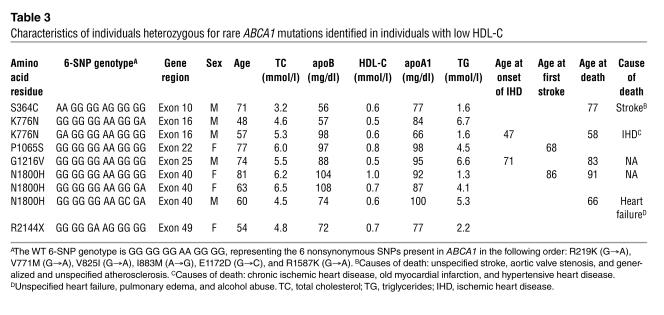
In the present study, 6 of the 7 mutations (except T774P) were identified only in individuals (n = 9) with low HDL-C (Table 2). Characteristics of these individuals are shown in Table 3. HDL-C levels ranged from 0.5 mmol/l to 1.0 mmol/l and apoAI levels from 66 mg/dl to 100 mg/dl. Triglycerides were increased (≥ 2.2 mmol/l) in 6 of 9 mutation carriers. Four unrelated individuals were heterozygous for mutations previously identified in Tangier disease (N1800H; n = 3) or in familial hypoalphalipoproteinemia (R2144X; n = 1). Four had known ischemic heart disease or stroke, 1 of which had died from chronic ischemic heart disease, and 2 had died recently of unknown causes. The cause of death in 2 other individuals was stroke and unspecified heart failure, respectively. Only 1 individual had premature ischemic heart disease, and none had clinical characteristics of Tangier disease.
Table 3.
Characteristics of individuals heterozygous for rare ABCA1 mutations identified in individuals with low HDL-C
The remaining 5 mutations in the coding region of ABCA1 did not introduce amino acid substitutions: A937, V990, E1211, and G2061 have not been identified previously, whereas V1053 has been reported by others (15) (Table 2).
Pairwise LD between SNPs.
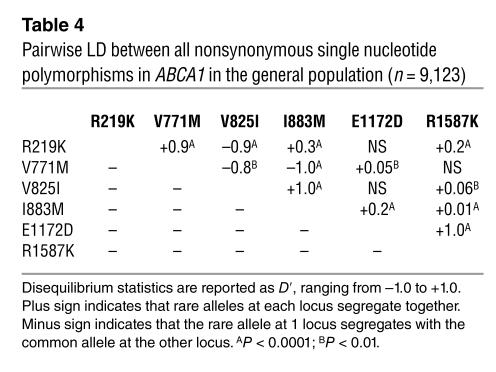
Pairwise LD performed for all 17 SNPs identified in individuals with low and high HDL-C revealed that LD between the promoter and the coding SNPs was very weak (Supplemental Table 3), most likely due to the fact that the human ABCA1 gene spans a genomic sequence of 150 kb. The nature of this LD structure justifies that estimation of haplotype frequencies is performed separately for the coding region. Strong pairwise LD was present in the coding region of ABCA1 for the following nonsynonymous SNP pairs: R219K with V771M (+LD) and V825I (–LD), V771M with V825I (–LD) and I883M (–LD), V825I with I883M (+LD), and E1172K with R1587K (+LD) (Table 4).
Table 4.
Pairwise LD between all nonsynonymous single nucleotide polymorphisms in ABCA1 in the general population (n = 9,123)
Haplotype analysis.
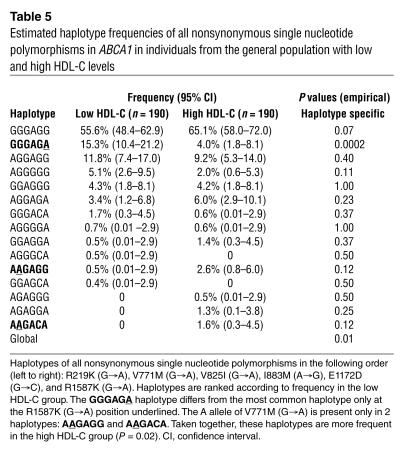
Haplotype analysis of the 6 nonsynonymous SNPs estimated that 7 of 17 haplotypes accounted for more than 97% of all haplotypes in the low HDL-C group and about 91% of all haplotypes in the high HDL-C group (Table 4). The global haplotype frequencies were significantly different between individuals with low and high HDL-C (P = 0.01). When specific haplotypes were examined, the unambiguous haplotype with the rare A allele at position 4759 (R1587K), which only differed at this position from the most common haplotype (GGGAGG), was significantly more frequent in the low HDL-C group than in the high HDL-C group (P = 0.0002), while the 2 haplotypes with the rare A allele at position 2310 (V771M), taken together, were less frequent in the low HDL-C group than in the high HDL-C group (P = 0.02) (Table 5).
Table 5.
Estimated haplotype frequencies of all nonsynonymous single nucleotide polymorphisms in ABCA1 in individuals from the general population with low and high HDL-C levels
Association of nonsynonymous SNPs with variation in lipid levels in the general population.
We genotyped the total general population sample for all 6 nonsynonymous SNPs in ABCA1. When corrected for multiple comparisons, genotype frequencies did not differ from those predicted by the Hardy-Weinberg equilibrium.
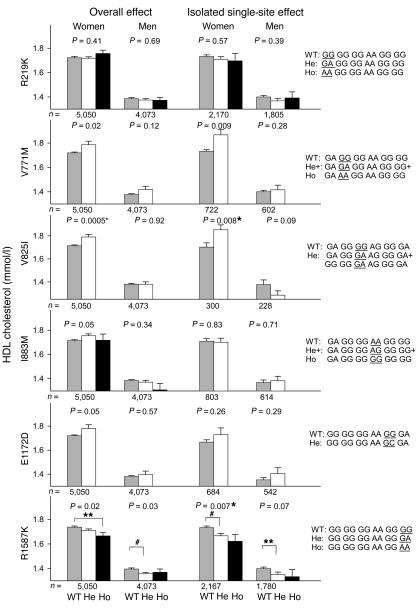
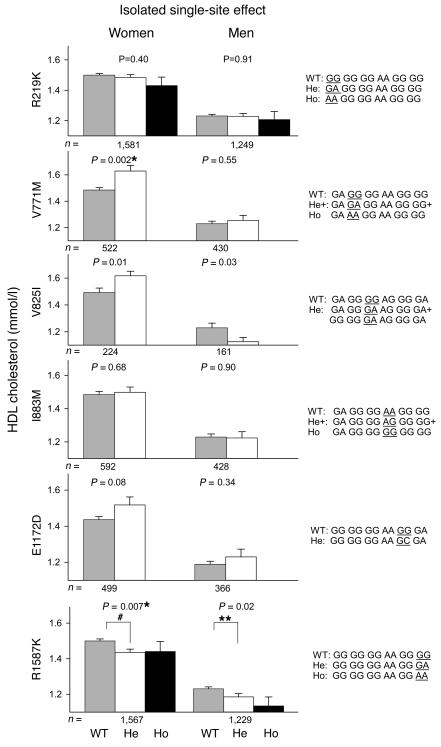
Overall effects (regardless of variation at the other 5 sites) for the 6 nonsynonymous SNPs on HDL-C levels are presented separately for each gender in Figure 4 (left panels). ANOVAs that fulfill a Bonferroni-corrected 2-sided significance level of P < 0.0083 were considered significant (marked with an asterisk in Figures 4–6). In women, V825I genotype was associated with significant increases in HDL-C of 0.08 mmol/l (GG versus GA+AA, P = 0.0005), and similar trends were found for V771M and E1172D (V771M: GG versus GA+AA, P = 0.02; E1172D: GG versus GC+CC, P = 0.05) (Figure 4, left panels). Due to the relatively low number of homozygous mutants, heterozygotes and homozygotes were pooled for some SNPs. In both women and men, R1587K genotype tended to decrease HDL-C (women: ANOVA: P = 0.02, post hoc test: GG versus AA, P = 0.02, ØHDL-C = –0.07 mmol/l; men: ANOVA: P = 0.03, post hoc test: GG versus GA, P = 0.008, ØHDL-C = –0.04 mmol/l) (Figure 4, left panels).
Figure 4.
In left panels, plasma HDL-C (mmol/l) as a function of 6 nonsynonymous SNPs in ABCA1 regardless of variation at the other 5 sites (overall effects) in women and men from the general population. In right panels plasma HDL-C (mmol/l) as a function of 6 SNP genotypes differing only at the SNP of interest (isolated single site effects) and in the order (left to right) R219K, V771M, V825I, I883M, E1172D, R1587K in women and men from the general population. The WT (the most common genotype), heterozygote (He), and homozygote (Ho) 6 SNP genotypes are given to the right of the bar graphs with the relevant SNP underlined. To increase power for V825I, the combined genotypes also varied at the R219K site, since R219K did not affect HDL-C levels. Values are mean ± SEM. *P < 0.0083 for all ANOVAs when corrected for multiple comparisons using the Bonferroni method. Post hoc tests: **P < 0.05; #P < 0.01.
Figure 6.
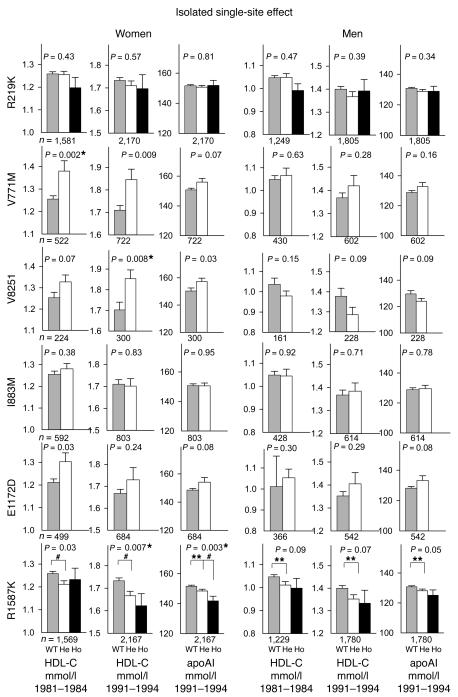
Plasma HDL-C (mmol/l) as a function of 6 SNP genotypes differing only at the relevant SNP (isolated single-site effects) as detailed in the legend to Figure 4, right panels. Values are estimated marginal mean ± SE for 2 independent measurements of plasma HDL-C obtained from the same individuals at 2 different time points corresponding to the HDL-C levels shown individually in Figure 5, respectively, for the second (1981–1984) and third examinations (1991–1994) of the Copenhagen City Heart Study. Analyses by repeated measures ANOVA. *P < 0.0083 for all ANOVAs when corrected for multiple comparisons using the Bonferroni method. Post hoc tests: **P < 0.05; #P < 0.01.
To ensure that the above effects were not dependent on LD with other SNPs, we tested the isolated single site effect of each specific SNP by comparing the 6 SNP genotypes that differed only at the SNP of interest, or for V825I at the SNP of interest and at the R219K site, since R219K did not affect HDL-C levels (Figure 4, right panels). Using this approach, the analyses became more accurate, although the number of individuals included decreased substantially, between 2-fold and 16-fold. In women, V825I was associated with increases in HDL-C of 0.15 mmol/l (V825I: GG versus GA, P = 0.008), with a similar trend for V771M (GG versus GA+AA, P = 0.009) (Figure 4, right panels). R1587K was associated with a stepwise decrease in HDL-C in women of 0.07 mmol/l and 0.11 mmol/l in heterozygotes and homozygotes, respectively (ANOVA: P = 0.007; post hoc tests: GG versus GA, P = 0.006, and GG versus AA, P = 0.08), and with a similar trend in men (ANOVA: P = 0.07; post hoc test: GG versus GA, P = 0.04, ØHDL-C= –0.05 mmol/l) (Figure 4, right panels).
To further verify these effects on HDL-C, we retested all 6 nonsynonymous SNPs on HDL-C measured 10 years earlier at the second examination of the Copenhagen City Heart Study (1981–1984). Figure 5 shows the isolated single site effects of 6 SNP genotypes corresponding to Figure 4 (right panels), but now further extended with HDL-C from the 1981–1984 examination and with apoAI measurements from the 1991–1994 examination. Figure 5 emphasizes that in women V771M and V825I are associated with increases in HDL-C and with similar trends in apoAI, while R1587K is associated with stepwise decreases in HDL-C and apoAI in women, and with similar trends in men. These associations were all consistent over time although P values changed slightly.
Figure 5.
Plasma HDL-C (mmol/l) measured at the second examination (1981–1984) and HDL-C and apoAI measured at the third examination (1991–1994) of the Copenhagen City Heart Study as a function of 6 SNP genotypes differing only at the relevant SNP (isolated single site effects) as detailed in the legend to Figure 4, right panels. Values are mean ± SEM. *P < 0.0083 for all ANOVAs when corrected for multiple comparisons using the Bonferroni method. Post hoc tests: **P < 0.05; #P < 0.01.
To ensure that optimal statistical power was gained from testing samples obtained from the same individuals at independent time points, repeated measures ANOVA was performed on the 2 measurements of HDL-C from the second (1981–1984) and third (1991–1994) examinations, respectively, of the Copenhagen City Heart Study (Figure 6). Estimated marginal means for the 2 HDL-C measurements as a function of SNP genotypes (isolated single site effects) are presented in Figure 6. The data show essentially the same as Figure 4 (right panels) and Figure 5, although the number of individuals in the analyses were substantially reduced to those who participated in both the second and third examinations.
In contrast, there were no consistent, overall, isolated single site or time consistent effects on HDL-C for R219K, I883M, or E1172D in either sex (Figures 4–6). Finally, none of the 6 nonsynonymous SNPs examined were associated with variation in levels of total cholesterol, triglycerides, or apoB (data not shown).
Discussion
With the aim to identify genetic variation in ABCA1 affecting HDL-C levels, we used a systematic approach in which we screened the core promoter and all 50 exons of ABCA1 in 190 individuals with extreme HDL-C levels selected from a large sample of the general population (n = 9,259). We subsequently genotyped the entire general population sample for the 6 nonsynonymous SNPs identified, and determined the effect of these SNPs on lipid, lipoprotein, and apolipoprotein levels as single sites and as combined genotypes differing only at the relevant SNP. Finally, we verified the results at 2 different examinations 10 years apart.
Novel observations in this study include the following: (a) we identified 3 new missense mutations that the evidence strongly suggests are associated with hypoalphalipoproteinemia; (b) we showed that 4 of 9 individuals heterozygous for mutations associated with low HDL-C carry 1 or the other of 2 mutations previously identified in families with Tangier disease or familial hypoalphalipoproteinemia; (c) we demonstrated that 10% (9 of 95) of individuals in the general population with low HDL-C are heterozygous for mutations in ABCA1; (d) we showed that the minimum frequency of rare mutations in ABCA1 associated with hypoalphalipoproteinemia in the general population is about 1 in 1,000, or considerably higher than previously assumed; (e) we demonstrated that common nonsynonymous SNPs and haplotypes harboring these SNPs may segregate differently in individuals from the general population with low and high HDL-C levels; (f) and finally, we demonstrated that 3 of 6 nonsynonymous SNPs affect HDL-C and apoAI levels in the general population and that these results were consistent over time.
SNPs in coding regions.
The present systematic screening approach of individuals with extreme HDL-C levels has proven to be sensitive not only in detecting ABCA1 mutations with strong functional consequence, but also in the detection of SNPs that affect HDL-C levels, illustrated by differential segregation of some of the functional SNPs in groups with extreme phenotypes. These findings are supported by previous observations for SNPs in the APOAV gene affecting levels of triglycerides (23). Whether one can pick up such a frequency difference between extreme phenotype groups depends, however, on the size of the study, the frequency of the SNP, the order of magnitude of the phenotype effect of the SNP (in casu on HDL-C), and whether this effect is equally strong in both genders.
We detected all previously reported SNPs in the coding region and core promoter of the gene as well as a new synonymous SNP (T1512) and ten new SNPs in introns. The T1512 SNP was the least frequent of the 12 SNPs identified in the coding region, suggesting that we did not overlook any important SNPs.
In women in the general population, the 2 relatively rare SNPs, V771M (0.03) and V825I (0.06), were both associated with increases in HDL-C of similar magnitude, which were consistent over time. The common SNP, R1587K (0.24), was associated with stepwise decreases in HDL-C and apoAI in women and were consistent over time, with similar trends in men. A recent report supports that R1587K affects levels of apoAI (14). Taken together, this suggests that the lack of significant association in men for V771M and V825I was partly due to less-significant effects on HDL-C in men of ABCA1 SNPs in general, in combination with the very low frequency of these 2 SNPs compared with R1587K. The frequencies of the remaining SNPs (R219K, I883M, E1172D) did not differ between individuals with low and high HDL-C and did not show time-consistent effects on HDL-C. Thus, although the rare A alleles of V771M and R1587K were both in LD with the rare A and C alleles of R219K and E1172D, the high and low HDL-C group frequencies, the haplotype data, the isolated single site effects of the SNPs, and the time-consistent phenotypic effects clearly show that only V771M and R1587K have independent effects on HDL-C levels.
Associations between all identified nonsynonymous ABCA1 SNPs and lipid traits have not been reported previously in a large sample representative of the general population. The initial reports on the R219K SNP suggested decreased atherosclerosis and age-dependent effects on cholesterol efflux of the K variant (15), but failed to show an effect on HDL-C levels. In agreement with this, we and others (14) do not find an association between the R219K SNP and HDL-C. We further document this for HDL-C levels measured 10 years earlier and for apoAI levels both as overall and isolated single site effects. Because the early previous studies (15–17) have not taken all pairwise LDs of nonsynonymous SNPs throughout the coding part of ABCA1 into account, these studies cannot determine whether the observed effects are independent or due to LD with other SNPs.
Since the error problem associated with assigning an estimated haplotype to a phenotype (in reality the result of 2 haplotypes) in haplotype-phenotype algorithms has not been solved (14), the present approach to compare 6 SNP genotypes that differ only at the SNP of interest is a simple way of illustrating which sites are functional and which are not. As illustrated in the present study, however, this approach requires a very large sample size.
Despite the large number of subjects analyzed, the biological effect of the nonsynonymous ABCA1 SNPs on the intermediate phenotype, HDL-C levels, is relatively modest, as would be expected for common variants. Thus, most of the genetics contributing to extreme HDL-C levels in the general population are due to rare mutations with large effects in several different HDL-C genes, to interactions between SNPs in 1 or more genes, or between SNPs and environmental factors (such as sex).
Mutations in coding regions.
We identified 3 new mutations (S364C, P1065S, G1216V) in individuals with low HDL-C that most likely cause familial hypoalphalipoproteinemia due to 3 important observations: location in important functional domains (18, 19, 24); location in highly conserved regions as well as total conservation across species at the exact residue; and location of previously reported disease-causing mutations in close vicinity to and/or of similar amino acid changing properties (2–4, 16, 20, 21, 25–29). The consequence of the S364C mutation is likely to be alteration of protein conformation caused by the introduction of a cysteine in the N-terminal extracellular loop prone to pair and introduce additional disulfide bonding (24). The P1065S mutation most likely interferes with normal ATP-binding in the first ABC: CFTR and ABCR/ABCA4 missense mutations in the corresponding regions are know to cause cystic fibrosis and Stargardt disease (30, 31). G1216V disrupts a stretch of highly hydrophilic amino acids in the R1 segment of the large central regulatory region. Furthermore, the new mutations, S364C, P1065S, and G1216V, were not identified in a random sample of approximately 1,500 individuals (3,000 alleles) from the general population, suggesting that they are indeed rare and restricted to individuals with low HDL-C.
We identified 4 previously reported mutations (T774P, K776N, N1800H, R2144X). The K776N mutation — in contrast to T774P — was identified only in individuals with low HDL-C (n = 2), was completely conserved between species, and was relatively conserved between paralogous human proteins. In the corresponding region of the CFTR/ABCC7 gene an R347P mutation (corresponding to residue 764 in ABCA1) segregates with cystic fibrosis, and this mutation also substitutes a polar residue for an uncharged amino acid (32). The N1800H and R2144X mutations that were present in 4 of 9 mutation carriers in the low HDL-C group have been reported previously in Tangier disease and in familial hypoalphalipoproteinemia, respectively (20–22). This suggests that mutations in ABCA1 found in families with Tangier disease or familial hypoalphalipoproteinemia — in the heterozygote state — may be relatively common causes of hypoalphalipoprotenemia in the general population. In the present study, we have identified 3 unrelated heterozygous carriers of the N1800H mutation among the 95 individuals with low HDL-C, suggesting that this variant is a potential founder mutation in Denmark.
The characteristics of the 9 mutation carriers illustrate that the lipid phenotype of hypoalphalipoproteinemia in the general population is heterogeneous. We chose initially, after careful consideration, to select extreme HDL-C groups for the ABCA1 screening solely based on HDL-C levels adjusted for sex and age. Had we designed a set of selection criteria, that is, normal triglyceride levels, we would not have detected 6 of the 9 probands.
Because not all of the ABCA1 gene was screened in individuals with high HDL-C, there could be additional ABCA1 mutations in high HDL-C subjects that were not detected with the current screening protocol. In this group we screened only 28 of 51 ABCA1 fragments, namely those fragments in which mutations and SNPs had been identified in the low HDL-C group.
The present systematic screening of ABCA1 suggests that at least 10% of individuals with low HDL-C in the general population are heterozygous for mutations in ABCA1 and that both mutations and SNPs in ABCA1 contribute to variation in HDL-C and apoAI levels in the general population.
Methods
Subjects.
The Copenhagen City Heart Study is a prospective cardiovascular population study of individuals selected based on the Central Population Register Code to reflect the adult Danish general population aged 20–80+ years. At the third examination, 1991–1994, 9,259 participants (55% women) gave blood for DNA analyses. More than 99% were white and of Danish descent (33, 34). All participants gave written informed consent, and the study was approved by the local ethical committee: number 100.2039/91, Copenhagen and Frederiksberg Committee.
For the genetic screening of ABCA1, we selected individuals from The Copenhagen City Heart Study with the lowest 1% (n = 95) and highest 1% (n = 95) HDL-C levels for age (in 10-year age groups) and sex. Consequently, the cut-off levels for HDL-C depend upon the 7 age groups for each sex. For the low HDL-C group (women and men, respectively) the levels are as follows: 20–29 years, 1.0 and 0.8 mmol/l; 30–39 years, 0.9 and 0.7 mmol/l; 40–49 years, 0.8 and 0.6 mmol/l; 50–59 years, 0.7 and 0.6 mmol/l; 60–69 years, 0.7 and 0.7 mmol/l; 70–79 years, 0.8 and 0.6 mmol/l; and 80+ years, 1.0 and 0.8 mmol/l. For the high HDL-C group the levels are as follows: 20–29 years, 2.6 and 2.1 mmol/l; 30–39 years, 2.8 and 2.2 mmol/l; 40–49 years, 3.0 and 2.5 mmol/l; 50–59 years, 3.4 and 2.9 mmol/l; 60–69 years, 3.5 and 3.2 mmol/l; 70–79 years, 3.6 and 2.9 mmol/l; and 80+ years, 2.7 and 2.3 mmol/l. By screening these groups with extreme phenotypes, we increased the likelihood of identifying mutations and SNPs (rare allele frequency ≤1% and >1%, respectively) with impact on HDL-C levels in the general population. Initially, the core promoter and all 50 exons, including exon-intron boundaries of ABCA1, were screened for genetic variation in individuals with low HDL-C, and variants identified in regulatory and transcribed parts of the gene (in the core promoter and in exons 1–10, 15–19, 21–22, 24–25, 27–28, 31–32, 35, 40, 46, 49) in this group were subsequently screened in individuals with high HDL-C. Because not all of the ABCA1 gene was screened in individuals with high HDL-C, there could be additional ABCA1 mutations in high HDL-C subjects that were not detected with the current screening protocol.
All 6 nonsynonymous SNPs identified by screening ABCA1 were genotyped in the entire general population sample (n = 9,259), and the effect of each SNP on variation in lipid, lipoprotein, and apolipoprotein levels was determined as overall effects (regardless of variation at the other 5 sites) and as isolated single site effects (6 SNP genotypes differing only at the relevant SNP), using data from the third examination of the Copenhagen City Heart Study (1991–1994). These results were then verified using data from the second examination of the Copenhagen City Heart Study 10 years earlier (1981–1984).
In tables and figures, data for the general population sample are presented only for those individuals (n = 9,123 out of n = 9,259) where the 6 SNP genotypes for all nonsynonymous SNPs were available.
Gene screening.
Genomic DNA was isolated from frozen whole blood (QiaAmp4 DNA Blood Mini Kit; QIAGEN GmbH). Fifty-one PCR fragments were amplified covering 209 bp of the promoter region (core promoter), all 50 exons of ABCA1, and exon-intron boundaries. Primer sequences are available on-line (Supplemental Table 1), and PCR conditions are available from the authors. Mutational analysis of the PCR products was performed by denaturing HPLC (dHPLC), using the Wave DNA Fragment Analysis System (Transgenomic Inc.). The dHPLC buffers and run conditions are available from the authors. PCR fragments showing heteroduplex formation by dHPLC were subsequently sequenced on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems Inc.).
SNP genotyping in the general population.
The ABI PRISM 7900HT Sequence Detection System (Applied Biosystems Inc.) was used to genotype the general population sample for all nonsynonymous SNPs identified. TaqMan-based assays were used (probe and primer sequences available from the authors). Combined genotypes for all nonsynonymous SNPs were available on 9,123 individuals of the total cohort of 9,259 individuals.
Biochemical analyses.
Colorimetric and turbidimetric assays (Hitachi autoanalyzer) were used to measure plasma levels of total cholesterol, HDL-C, triglycerides, and apoB and apoAI (all from Boehringer Mannheim GmbH).
Statistical analyses.
Differences in allele frequencies between individuals with low and high HDL-C were evaluated using Fischer’s exact test. Pairwise disequilibrium statistics were calculated from D = h – pq, where h is the frequency of the rare estimated haplotype for a pair of sites and p and q are the frequencies, assuming no linkage, of the alleles in that haplotype (35). LD was expressed as D′ (36). Significance levels for pairwise LD were estimated by the likelihood-ratio test. Estimated haplotype frequencies were calculated using the expectation maximization algorithm (37) (http://linkage.rockefeller.edu/ott/eh.htm). The effect of genotype on variation in lipid, lipoprotein, and apo levels was determined by ANOVA. Repeated measures ANOVA was applied to test the effect of SNPs as isolated single sites on HDL-C levels in samples obtained from the same individuals at 2 independent time points 10 years apart at the second (1981–1984) and third (1991–1994) examinations of the Copenhagen City Heart Study, respectively. A Bonferroni corrected P value less than 0.0083 on a 2-sided test (6 different nonsynonymous SNPs tested) was considered significant.
Supplementary Material
Acknowledgments
We thank Christina Dam and Mette Refstrup for expert technical assistance. We also thank the subjects who participated in the study. This work was supported by The Danish Heart Foundation, The Danish Medical Research Council, Ingeborg and Leo Dannin’s Grant, and the Research Fund at Rigshospitalet, Copenhagen University Hospital.
Footnotes
See the related Commentary beginning on page 1244.
Nonstandard abbreviations used: ABCA1, ABC transporter A1; dHPLC, denaturing high-performance liquid chromatography; HDL-C, HDL cholesterol; LD, linkage disequilibrium; SNP, single-nucleotide polymorphism.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Assman, G., von Eckardstein, A., and Bryan Brewer, Jr., H. 2001. Familial analphalipoproteinemia: Tangier disease. In The metabolic and molecular bases of inherited disease. 8th edition. C.R. Scriver, A.L. Beaudet, W.S. Sly, and D. Valle, editors. McGraw-Hill. New York, New York, USA. 2937–2960.
- 2.Bodzioch M, et al. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat. Genet. 1999;22:347–351. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- 3.Brooks-Wilson A, et al. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat. Genet. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- 4.Rust S, et al. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat. Genet. 1999;22:352–355. doi: 10.1038/11921. [DOI] [PubMed] [Google Scholar]
- 5.Schmitz G, Langmann T. Structure, function and regulation of the ABC1 gene product. Curr. Opin. Lipidol. 2001;12:129–140. doi: 10.1097/00041433-200104000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz G, Kaminski WE, Orso E. ABC transporters in cellular lipid trafficking. Curr. Opin. Lipidol. 2000;11:493–501. doi: 10.1097/00041433-200010000-00007. [DOI] [PubMed] [Google Scholar]
- 7.Dean M, Hamon Y, Chimini G. The human ATP-binding cassette (ABC) transporter superfamily. J. Lipid. Res. 2001;42:1007–1017. [PubMed] [Google Scholar]
- 8.Friedlander Y, Kark JD, Stein Y. Biological and environmental sources of variation in plasma lipids and lipoproteins: the Jerusalem Lipid Research Clinic. Hum. Hered. 1986;36:143–153. doi: 10.1159/000153618. [DOI] [PubMed] [Google Scholar]
- 9.Hunt SC, et al. Genetic heritability and common environmental components of resting and stressed blood pressures, lipids, and body mass index in Utah pedigrees and twins. Am. J. Epidemiol. 1989;129:625–638. doi: 10.1093/oxfordjournals.aje.a115175. [DOI] [PubMed] [Google Scholar]
- 10.Tall, A.R., Breslow, J.L., and Rubin, E.M. 2001. Genetic disorders affecting plasma high-density lipoproteins. In The metabolic and molecular bases of inherited disease. 8th edition. C.R. Scriver, A.L. Beaudet, W.S. Sly, and D. Valle, editors. McGraw-Hill. New York, New York, USA. 2915–2936.
- 11.Pullinger CR, et al. Analysis of hABC1 gene 5′ end: additional peptide sequence, promoter region, and four polymorphisms. Biochem. Biophys. Res. Commun. 2000;271:451–455. doi: 10.1006/bbrc.2000.2652. [DOI] [PubMed] [Google Scholar]
- 12.Zwarts KY, et al. ABCA1 regulatory variants influence coronary artery disease independent of effects on plasma lipid levels. Clin. Genet. 2002;61:115–125. doi: 10.1034/j.1399-0004.2002.610206.x. [DOI] [PubMed] [Google Scholar]
- 13.Iida A, et al. High-density single-nucleotide polymorphism (SNP) map of the 150-kb region corresponding to the human ATP-binding cassette transporter A1 (ABCA1) gene. J. Hum. Genet. 2001;46:522–528. doi: 10.1007/s100380170034. [DOI] [PubMed] [Google Scholar]
- 14.Treqouet D-A, et al. In-depth haplotype analysis of ABCA1 gene polymorphisms in relation to plasma apoAI levels and myocardial infarction. Arterioscler. Thromb. Vasc. Biol. 2004;24:775–781. doi: 10.1161/01.ATV.0000121573.29550.1a. [DOI] [PubMed] [Google Scholar]
- 15.Clee SM, et al. Common genetic variation in ABCA1 is associated with altered lipoprotein levels and a modified risk for coronary artery disease. Circulation. 2001;103:1198–1205. doi: 10.1161/01.cir.103.9.1198. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, et al. Common and rare ABCA1 variants affecting plasma HDL-C. Arterioscler. Thromb. Vasc. Biol. 2000;20:1983–1989. doi: 10.1161/01.atv.20.8.1983. [DOI] [PubMed] [Google Scholar]
- 17.Brousseau ME, et al. Common variants in the gene encoding ATP-binding cassette transporter 1 in men with low HDL-C levels and coronary heart disease. Atherosclerosis. 2001;154:607–611. doi: 10.1016/s0021-9150(00)00722-x. [DOI] [PubMed] [Google Scholar]
- 18.Fitzgerald ML, et al. Naturally occurring mutations in the largest extracellular loops of ABCA1 can disrupt its direct interaction with apolipoprotein A-I. J. Biol. Chem. 2002;277:33178–33187. doi: 10.1074/jbc.M204996200. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka AR, et al. Effects of mutations of ABCA1 in the first extracellular domain on subcellular trafficking and ATP-binding/hydrolysis. J. Biol. Chem. 2003;278:8815–8819. doi: 10.1074/jbc.M206885200. [DOI] [PubMed] [Google Scholar]
- 20.Clee SM, et al. Age and residual cholesterol efflux affect HDL-C levels and coronary artery disease in ABCA1 heterozygotes. J. Clin. Invest. 2000;106:1263–1270. doi: 10.1172/JCI10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brousseau ME, et al. Novel mutations in the gene encoding ATP-binding cassette 1 in four Tangier disease kindreds. J. Lipid. Res. 2000;41:433–441. [PubMed] [Google Scholar]
- 22.Marcil M, et al. Mutations in the ABC1 gene in familial HDL deficiency with defective cholesterol efflux. Lancet. 1999;354:1341–1346. doi: 10.1016/s0140-6736(99)07026-9. [DOI] [PubMed] [Google Scholar]
- 23.Pennacchio LA, et al. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 2001;294:169–173. doi: 10.1126/science.1064852. [DOI] [PubMed] [Google Scholar]
- 24.Bungert S, Molday LL, Molday RS. Membrane topology of the ATP-binding cassette transporter ABCR and its relationship to ABC1 and related ABCA transporters: identification of N-linked glycosylation sites. J. Biol. Chem. 2001;276:23539–23546. doi: 10.1074/jbc.M101902200. [DOI] [PubMed] [Google Scholar]
- 25.Lapicka-Bodzioch K, et al. Homogeneous assay based on 52 primer sets to scan for mutations of the ABCA1 gene and its application in genetic analysis of a new patient with familial high-density lipoprotein deficiency syndrome. . Biochim. Biophys. Acta. 2001; 1537:42–48. doi: 10.1016/s0925-4439(01)00053-9. [DOI] [PubMed] [Google Scholar]
- 26.Huang W, et al. Novel mutations in ABCA1 gene in Japanese patients with Tangier disease and familial high density lipoprotein deficiency with coronary heart disease. . Biochim. Biophys. Acta. 2001; 1537:71–78. doi: 10.1016/s0925-4439(01)00058-8. [DOI] [PubMed] [Google Scholar]
- 27.Mott S, et al. Decreased cellular cholesterol efflux is a common cause of familial hypoalphalipoproteinemia: role of the ABCA1 gene mutations. Atherosclerosis. 2000;152:457–468. doi: 10.1016/s0021-9150(99)00498-0. [DOI] [PubMed] [Google Scholar]
- 28.Bertolini S, et al. A point mutation in ABC1 gene in a patient with severe premature coronary heart disease and mild clinical phenotype of Tangier disease. Atherosclerosis. 2001;154:599–605. doi: 10.1016/s0021-9150(00)00587-6. [DOI] [PubMed] [Google Scholar]
- 29.Miller M, Rhyne J, Hamlette S, Birnbaum J, Rodriguez A. Genetics of HDL regulation in humans. Curr. Opin. Lipidol. 2003;14:273–279. doi: 10.1097/00041433-200306000-00007. [DOI] [PubMed] [Google Scholar]
- 30.Karpowich N, et al. Crystal structures of the MJ1267 ATP-binding cassette reveal an induced-fit effect at the ATPase active site of an ABC transporter. Structure (Camb.). 2001;9:571–586. doi: 10.1016/s0969-2126(01)00617-7. [DOI] [PubMed] [Google Scholar]
- 31.Allikmets R, et al. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat. Genet. 1997;15:236–246. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- 32.Dean M, et al. Multiple mutations in highly conserved residues are found in mildly affected cystic fibrosis patients. Cell. 1990;61:863–870. doi: 10.1016/0092-8674(90)90196-l. [DOI] [PubMed] [Google Scholar]
- 33.Appleyard M, Hansen AT, Jensen G, Schnohr P, Nyboe J. The Copenhagen City Heart Study. Østerbroundersøgelsen. A book of tables with data from the first examination (1976–78) and a five year follow-up (1981–83). The Copenhagen City Heart Study Group. Scand. J. Soc. Med. Suppl. 1989;41:1–160. [PubMed] [Google Scholar]
- 34.Schnohr P, Jensen G, Lange P, Scharling H, Appleyard M. The Copenhagen City Heart Study, Østerbroundersøgelsen, tables with data from the third examination 1991–1994. Eur. Heart J. Suppl. H. 2001;3:1–83. [Google Scholar]
- 35.Pericak-Vance, M.A. 1998. Linkage disequilibrium and allelic association. In Approaches to gene mapping in complex human diseases. J.L. Haines and M.A. Pericak-Vance, editors. John Wiley & Sons, Inc. New York, New York, USA. 323–333.
- 36.Thompson EA, Deeb S, Walker D, Motulsky AG. The detection of linkage disequilibrium between closely linked markers: RFLPs at the AI-CIII apolipoprotein genes. Am. J. Hum. Genet. 1988;42:113–124. [PMC free article] [PubMed] [Google Scholar]
- 37.Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol. Biol. Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- 38.Schneider E, Hunke S. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 39.den Dunnen JT, Antonarakis E. Nomenclature for the description of human sequence variations. Hum. Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- 40.National Center for Biotechnology Information. Single Nucleotide Polymorphism database (dbSNP). http://www.ncbi.nlm.nih.gov/SNP/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.