| NCBI National Center for Biotechnology Information |  |
3C74:
X-ray structure of the uridine phosphorylase from salmonella typhimurium in complex with 2,2'-anhydrouridine at 2.38a resolution
| Biological unit 1: | hexameric | ||
| Source organism: | Salmonella enterica subsp. enterica serovar Typhimurium | ||
| Number of proteins: | 6 (Uridine phosphorylase ▼) Protein molecule
close
|
||
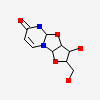
| Number of chemicals: | 6 (2,2'-Anhydro-(1-beta-D-arabinofuranosyl)uracil (6) ▼) Chemical
close |
| PDB ID | Description | Taxonomy | Aligned Protein | RMSD | Aligned Residues | Sequence Identity | |||
|---|---|---|---|---|---|---|---|---|---|
| 1 | Full |
1U1E | Structure of e. coli uridine phosphorylase complexed to 5(phenylseleno)acyclouridine (PSAU) |
Escherichia coli |
6 | 0.73Å | 1461 | 97% | |
| 2 | Full |
1U1D | Structure of e. coli uridine phosphorylase complexed to 5-(phenylthio)acyclouridine (ptau) |
Escherichia coli |
6 | 0.73Å | 1460 | 97% | |
| 3 | Full |
1RXS | E. coli uridine phosphorylase: 2'-deoxyuridine phosphate complex |
Escherichia coli |
6 | 0.59Å | 1458 | 97% | |
| 4 | Full |
1RXU | E. coli uridine phosphorylase: thymidine phosphate complex |
Escherichia coli |
6 | 0.66Å | 1458 | 97% | |
| 5 | Full |
2HRD | Crystal structure of the uridine phosphorylase from Salmonella typhimurium in complex with thymine and phosphate ion at 1.70A resolution |
Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 |
6 | 0.78Å | 1457 | 100% | |
| 6 | Full |
1U1C | Structure of E. coli uridine phosphorylase complexed to 5-benzylacyclouridine (BAU) |
Escherichia coli |
6 | 0.74Å | 1456 | 97% | |
| 7 | Full |
2HN9 | Crystal structure of the uridine phosphorylase from Salmonella typhimurium in complex with thymine and phosphate ion at 2.12A resolution |
Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 |
6 | 0.50Å | 1455 | 100% | |
| 8 | Full |
1RXC | E. COLI uridine phosphorylase: 5-fluorouracil ribose-1-phosphate complex |
Escherichia coli |
6 | 0.62Å | 1454 | 97% | |
| 9 | Full |
1U1G | Structure of E. coli uridine phosphorylase complexed to 5-(m-(benzyloxy)benzyl)barbituric acid (BBBA) |
Escherichia coli |
6 | 0.69Å | 1452 | 97% | |
| 10 | Full |
4OGK | X-ray structure of the uridine phosphorylase from Salmonella typhimurium in complex with thymidine at 2.40 A resolution |
Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 |
6 | 0.94Å | 1452 | 100% |