| NCBI National Center for Biotechnology Information |  |
3DPS:
X-ray structure of the unliganded uridine phosphorylase from salmonella typhimurium in homodimeric form at 1.8A
| Biological unit 1: | hexameric | ||
| Source organism: | Salmonella enterica subsp. enterica serovar Typhimurium | ||
| Number of proteins: | 6 (Uridine phosphorylase ▼) Protein molecule
close
|
||
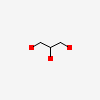
| Number of chemicals: | 3 (GLYCEROL (3) ▼) Chemical
close |
| PDB ID | Description | Taxonomy | Aligned Protein | RMSD | Aligned Residues | Sequence Identity | |||
|---|---|---|---|---|---|---|---|---|---|
| 1 | Full |
2OXF | X-ray structure of the unliganded uridine phosphorylase from Salmonella typhimurium in homodimeric form at 1.76A resolution |
Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 |
6 | 0.15Å | 1425 | 100% | |
| 2 | Full |
6RCA | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with 2.2'-anhydrouridine at 1.34 A |
Vibrio cholerae |
6 | 1.31Å | 1420 | 75% | |
| 3 | Full |
4I2V | X-ray structure of the unliganded uridine phosphorylase from Yersinia pseudotuberculosis at 2.12A resolution |
Yersinia pseudotuberculosis |
6 | 1.04Å | 1419 | 93% | |
| 4 | Full |
1RXY | E. coli uridine phosphorylase: type-B native |
Escherichia coli |
6 | 1.08Å | 1419 | 97% | |
| 5 | Full |
4NY1 | X-ray structure of the unliganded uridine phosphorylase from Yersinia pseudotuberculosis at 1.7 A resolution |
Yersinia pseudotuberculosis IP 32953 |
6 | 1.21Å | 1419 | 93% | |
| 6 | Full |
4R31 | Crystal structure of a putative uridine phosphorylase from Actinobacillus succinogenes 130Z (Target NYSGRC-029667 ) |
Actinobacillus succinogenes 130Z |
6 | 1.28Å | 1418 | 69% | |
| 7 | Full |
4OF4 | X-ray structure of unliganded uridine phosphorylase from Yersinia pseudotuberculosis at 1.4 A resolution |
Yersinia pseudotuberculosis |
6 | 1.19Å | 1416 | 93% | |
| 8 | Full |
1TGV | Structure of E. coli Uridine Phosphorylase complexed with 5-Fluorouridine and sulfate |
Escherichia coli |
6 | 1.27Å | 1416 | 97% | |
| 9 | Full |
1TGY | Structure of E. coli Uridine Phosphorylase complexed with uracil and ribose 1-phosphate |
Escherichia coli |
6 | 1.35Å | 1416 | 97% | |
| 10 | Full |
6EYP | X-ray structure of the unliganded uridine phosphorylase from Vibrio cholerae at 1.22A |
Vibrio cholerae |
6 | 1.37Å | 1416 | 76% |