Unravelling the therapeutic potential of forkhead box proteins in breast cancer: An update (Review)
- Authors:
- Published online on: June 4, 2024 https://doi.org/10.3892/or.2024.8751
- Article Number: 92
-
Copyright: © Anwar et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Breast cancer is a condition wherein the cells in the mammary gland grow in a dysregulated manner, resulting in the formation of a mass of cells called a tumor. The World Health Organization statistics revealed that, in 2020, the worldwide scale of breast cancer cases increased to 2.3 million, and, globally, 6,85,000 individuals lost their lives (1). Although breast cancer is generally considered benign, a delayed diagnosis can cause the cancer to spread to the surrounding lymph nodes, resulting in distant metastasis and impacting vital organs such as the brain, liver and lungs (2). Therefore, researchers are striving hard to develop effective therapeutic approaches; in this regard, the role of forkhead box proteins cannot be overlooked due to their involvement in cancer development and progression.
Forkhead box proteins, commonly known as forkhead box (FOX) proteins, belong to a family of transcription factors that are evolutionarily conserved, distinguished by their DNA binding domain, which is referred to as either ‘forkhead’ or ‘winged helix’ (3). These transcription factors play pivotal roles as essential regulators of gene expression in various pathological and physiological processes, encompassing cell proliferation, differentiation and survival (4). Interestingly, numerous studies have attempted to establish the links between FOX proteins and cellular events, including the cancer initiation and progression, alongside the emergence of resistance against anticancer drugs (5). These discoveries highlight the diverse functions of FOX proteins in the field of cancer biology.
The Forkhead box, or Fox, gene family is a group of transcriptional regulators known for their ancient evolutionary origins. They were named after the Drosophila melanogaster fork head gene (fkh). It has been reported that mutations in fkh can lead to abnormalities in head fold involution during embryonic development, causing adult flies to exhibit a characteristic spiked head phenotype (6). Fox genes are found across a vast range of organisms, from yeast to humans. They are further classified into subfamilies such as FOXA and FOXP. A defining feature of Fox proteins is their DNA-binding domain, the forkhead (FKH) region, which is remarkably similar across species and consists of roughly 100 amino acids. The high degree of conservation within the FKH domain extends across the entire Fox family. This feature served as the basis for the initial classification system, where Fox proteins were assigned to 15 classes, designated FOXA to FOXO, based on sequence similarity within the FKH domain. More recent evolutionary analyses have refined the Fox protein classification system, increasing the number of classes to 19, ranging from FOXP to FOXS. This categorization is based on sequence similarities within the FKH domain, which typically consists of three alpha helices, three beta sheets, and two unique ‘wing’ regions flanking the third beta sheet. The ‘forkhead’ or ‘winged helix’ terminology originates from the distinctive butterfly-like structure formed by these ‘wing’ regions in Fox proteins (7).
FOX transcription factors are crucial regulators in embryonic development and maintaining cellular balance, driven by evolutionary forces that shape their diverse functions (8). They act as master regulators of fundamental cellular functions, including cell cycle progression, proliferation, differentiation, DNA repair mechanisms, metabolism, blood vessel formation (angiogenesis), and ultimately, a cell's fate. Disruptions in Fox protein activity have been implicated in the onset, spread, advancement and resistance to therapy of cancer. They also influence pathways related to cancer, aiding cell survival in adverse conditions. The involvement of FOX proteins in cancer spans its entire spectrum, from initiation to metastasis, orchestrated through intricate networks. While the importance of Fox proteins is well-established, a comprehensive understanding of how mutations within FOX-binding sites in the regulatory regions of FOX-target genes remains incomplete. Further research is needed to unravel these mechanisms in detail.
FOX proteins can also be regulated by microRNAs (miRNAs), which are special non-coding RNAs whose role as a tumor suppressor and oncogene is gradually coming into stronger light (9,10). The present review has tried to provide a brief overview about miRNAs and FOX proteins as other multiple roles and association of FOX proteins have to be covered in more detail. However, a thorough analysis of the crosslinks between miRNAs and FOX proteins would help unlock the therapeutic hurdles around breast cancer treatment.
Currently, the understanding of the FOX family is at its nascent stage; therefore, a comprehensive study that unveils the complexity revolving around the FOX transcription factor and the detailed understanding of its association with breast cancer would aid in the discovery of cancer biomarkers and improve the existing treatment strategies. Keeping this in view, the article has tried to document the majority of important associations between multiple FOX proteins and breast cancer.
Role of various FOX proteins in breast cancer
The discovery of significant findings often sparks interest, and a similar scenario unfolded with FOX proteins, now recognized for their role in positive and negative regulation of breast carcinogenesis. Initially, the focus of the present review centered on the FOXA protein family, which is extensively engaged in hormonal signaling (10).
FOXA family and breast cancer
Considering the diverse functions and phenotypes of normal breast epithelial cells, gene expression profiling has facilitated the categorization of breast cancer into the following five types: Luminal type A, luminal type B, normal-like, ERα-negative, HER-2-positive and basal type (8). The luminal types A and B are characterized by ERα-positive due to the elevated expression of ERα, demonstrating an improved prognosis. FOXA1, a member of the forkhead family, is garnering attention for its significant involvement in the hormonal signaling network, which plays a crucial role in regulating the growth and differentiation of epithelial cells in breast cancer (10).
Hepatocyte nuclear factor 3α (HNF3α), also known as FOXA1, is identified as the first transcription factor of the FOXA family. It is enriched with hepatocytes and significantly regulates α1-antitrypsin gene and transthyretin expressions (11). Apart from FOXA1, the FOXA family comprises other members, namely FOXA2 (HNF3β) and FOXA3 (HNF3γ) (12). These transcriptional factors bind to ‘TGTTTAC’ or ‘TGTTTGC’ motifs present in the promoter/enhancer regions of the target genes (13). Each factor binds to the target gene with varying affinities, thereby modulating transcription. The FOXA factors are referred to as the ‘pioneer factors’ as they exhibit a unique capability to engage with target sites within silent chromatin. This distinctive ability enables them to initiate regulatory events, setting them apart from other transcription factors.
FOXA1 comprises a helix-turn-helix motif of 110 amino acids as well as three α-helices, two β-strands, and two polypeptide loops on either side, which are referred to as Wing1 and Wing2 subdomains (13,14). The DNA binding domain is equipped with a polypeptide chain on either side, giving it a wing-like appearance that is responsible for identifying the stability associated with DNA binding (12). The binding domain, along with the polypeptide chain, binds to the consensus sequence ‘A(A/T)TRTT(G/T)RYTY’ wherein the binding domain of the DNA interacts with the major grooves and the polypeptide chains interact with the minor grooves of DNA.
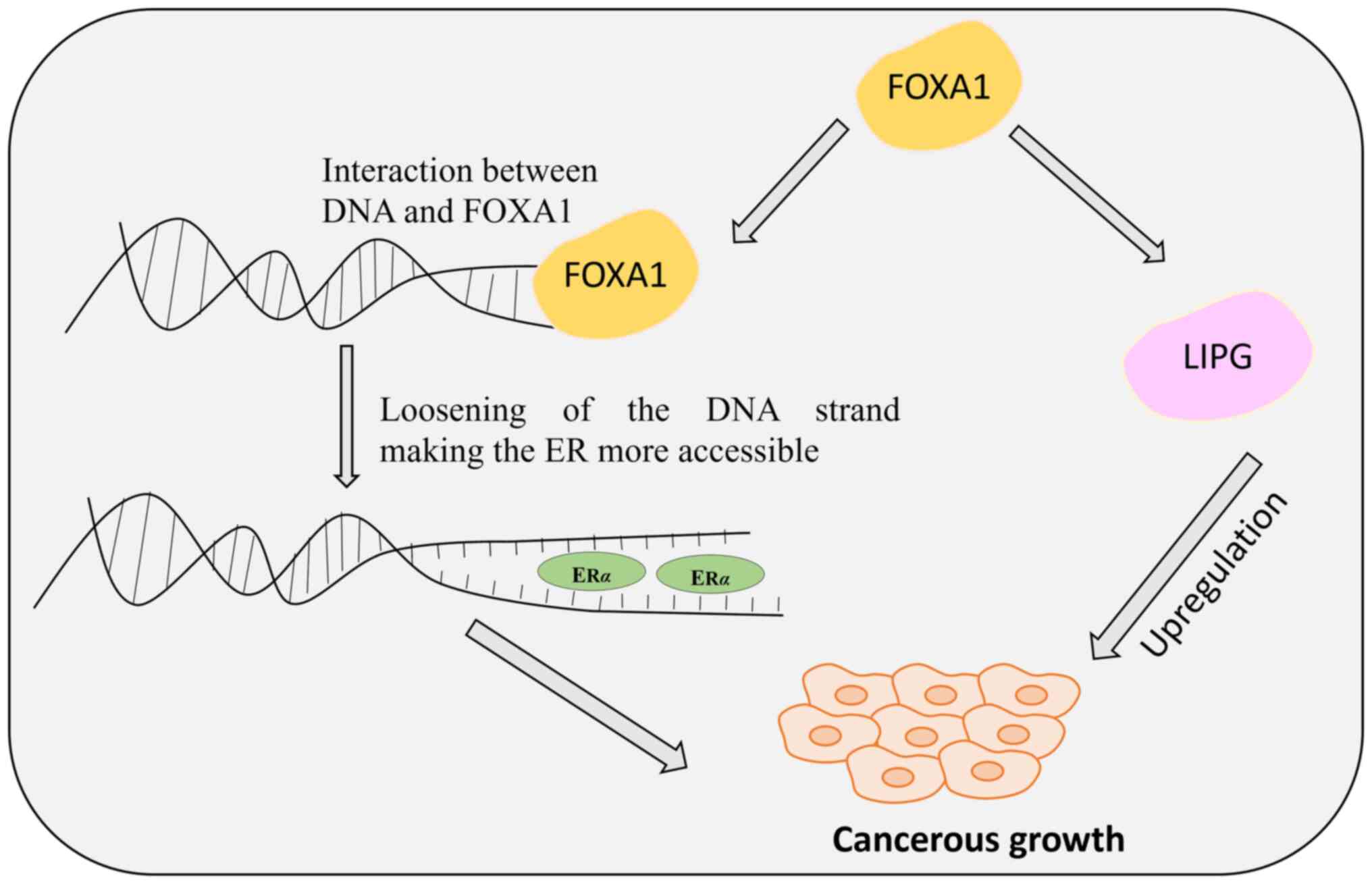
FOXA1 resembles linker histones in DNA binding and stabilizes the major groove by projecting into it, utilizing polypeptide loops similar to histones 1 and 5. Unlike the linker proteins, FOXA1 does not compact DNA around peptide sequences but enhances the nucleosome accessibility to facilitate interactions with transcriptional regulators (15,16). FOXA1 is vital for mammary morphogenesis and estrogen receptor (ER) activity and is pivotal in normal breast tissue development (17). In breast tissues, FOXA1 localizes within cell nuclei where it co-localizes with ER. FOXA1 interacts with specific DNA sequences termed as cis-regulatory elements, located within heterochromatin, thereby aiding in the binding of hormone receptors to chromatin. This ‘pioneering’ function opens up the chromatin structure, allowing other transcription factors, such as nuclear receptors, to bind and regulate gene expression (18). FOXA1 is crucial for various aspects of normal breast development, such as mammary gland morphogenesis (13), and is essential for optimal ER activity (19) (Fig. 1). Moreover, FOXA1 directly interacts with GATA3 (20), emphasizing its regulatory role in mammary gland development. Ghosh et al (21) proposed that hypermethylation in healthy breast tissue might reduce FOXA1 expression, potentially leading to impaired ER function and influencing breast tumor development. However, the precise role of FOXA1 in tumor initiation and progression remains a topic of ongoing investigation. It is widely accepted that FOXA1 depletion inhibits cancer cell proliferation (22), as both ER and androgen receptor transcriptional activities rely on FOXA1 in breast and prostate cancer cells. Additionally, during carcinogenesis, FOXA1 directly enhances the transcription of several downstream genes that support the luminal phenotype, such as the E-cadherin gene (CDH1) (23) and GATA3 (24), while simultaneously suppressing basal differentiation (25).
The lipid metabolism pathway is the primary pathway used by cancer cells to meet their energy requirements to proliferate (26). Apart from the ER levels, FOXA1 is known to directly regulate the endothelial lipase (LIPG) enzyme expression levels. LIPG, belonging to the lipoprotein lipase family, plays a substantial role in facilitating tumor growth by promoting breast cancer cell-lipid addiction, and its reduced expression causes tumor growth impairment. In breast cancer cells, the increased expression of FOXA1 elevates the levels of LIPG, which in turn stimulates tumor growth by enhancing cell proliferation (Fig. 1). LIPG works by generating lipid precursors extracellularly, which are transported to manage the intracellular lipid species (22). The downregulation of LIPG leads to impaired breast cancer cell growth, inhibition of proliferation, and tumor development.
FOXA1 serves dual role as a therapeutic target and a prognostic marker; however, further investigation is necessary to understand the relationship between FOXA1 levels in normal luminal breast tissue and the complex interactions between FOXA1, ER alpha (ERα), and LIPG expression. This knowledge gap hinders the ability to fully elucidate the mechanisms underlying breast cancer initiation and progression.
FOXO family and breast cancer
FOXOs, constitute another group within the family of forkhead proteins classified by a forkhead domain and a winged-helix DNA binding motif (27), have been identified as tumor suppressors, a key regulator in animal physiology, and control various cellular processes, such as cellular growth, differentiation, proliferation, apoptosis, DNA repair and stem cell maintenance.
FOXOs negatively control cancer progression by modulating angiogenesis and metastasis. At present, four members of FOXO family have been identified: FOXO1, FOXO3A, FOXO4 and FOXO6; each characterized by unique core functional domains (28). With the exception of FOXO6, the other three members have similar structures, whereas FOXO6 differs in structure as compared with the rest. However, despite the structural differences, all four members have the same DNA binding specificity and recognize the regulatory elements that contain the sequence ‘TTGTTTAC’. Additionally, all members are known to work with coactivators and corepressors to modulate their functioning.
Previous studies have demonstrated that, in cancer, FOXOs exist as chimeric products due to chromosomal translocation, and the presence of these hybrids has directed the attention of researchers toward the FOXOs' role in various cancers. FOXO1 is a fusion protein, with the partner being either PAX3 or PAX7 DBD in pediatric alveolar rhabdomyosarcoma (29). FOXO3A and FOXO4 were identified as fusion proteins with the mixed-lineage leukemia gene in acute myeloid leukemias (30). The existence of FOXOs as fusion proteins indicates their role in human cancers.
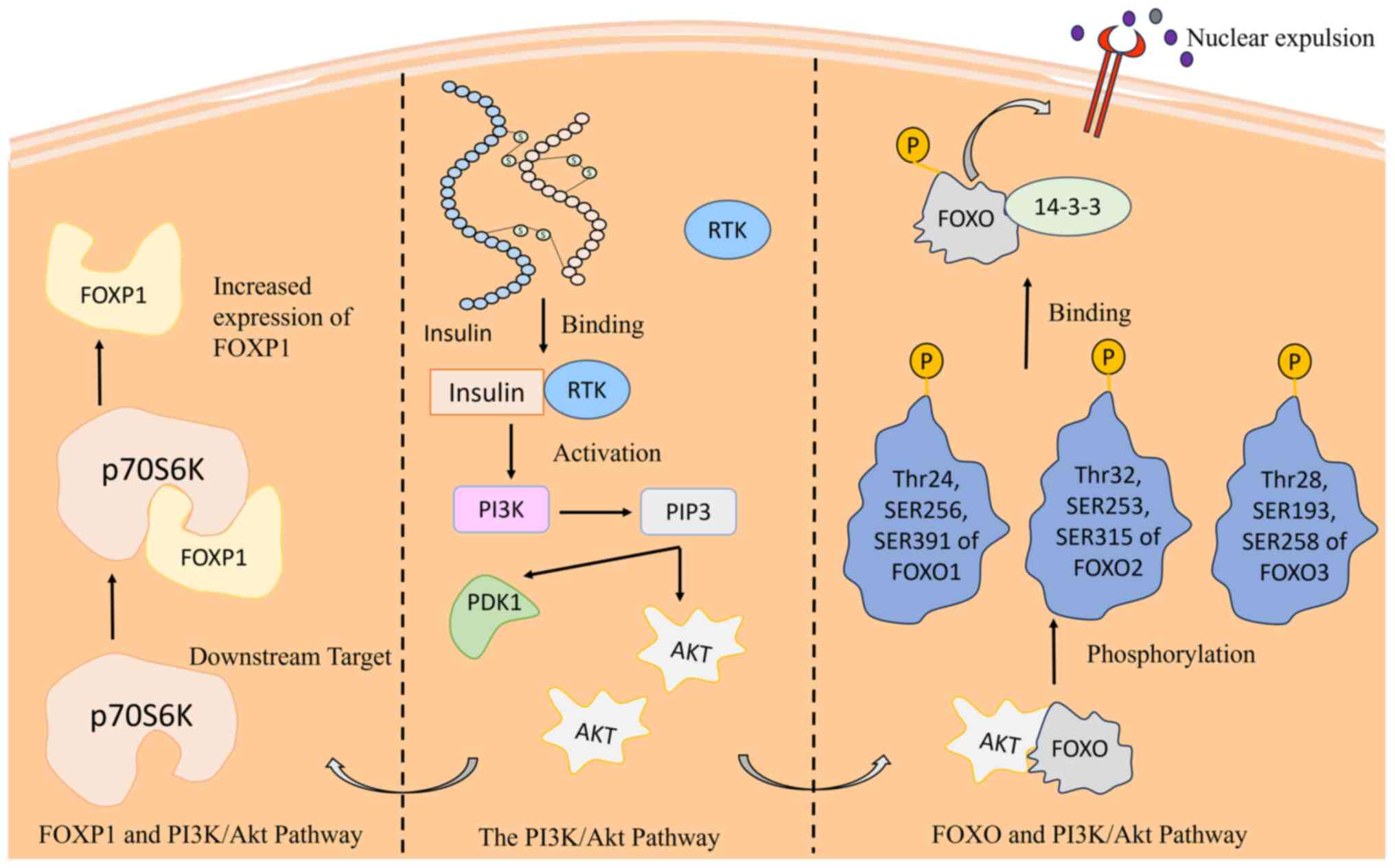
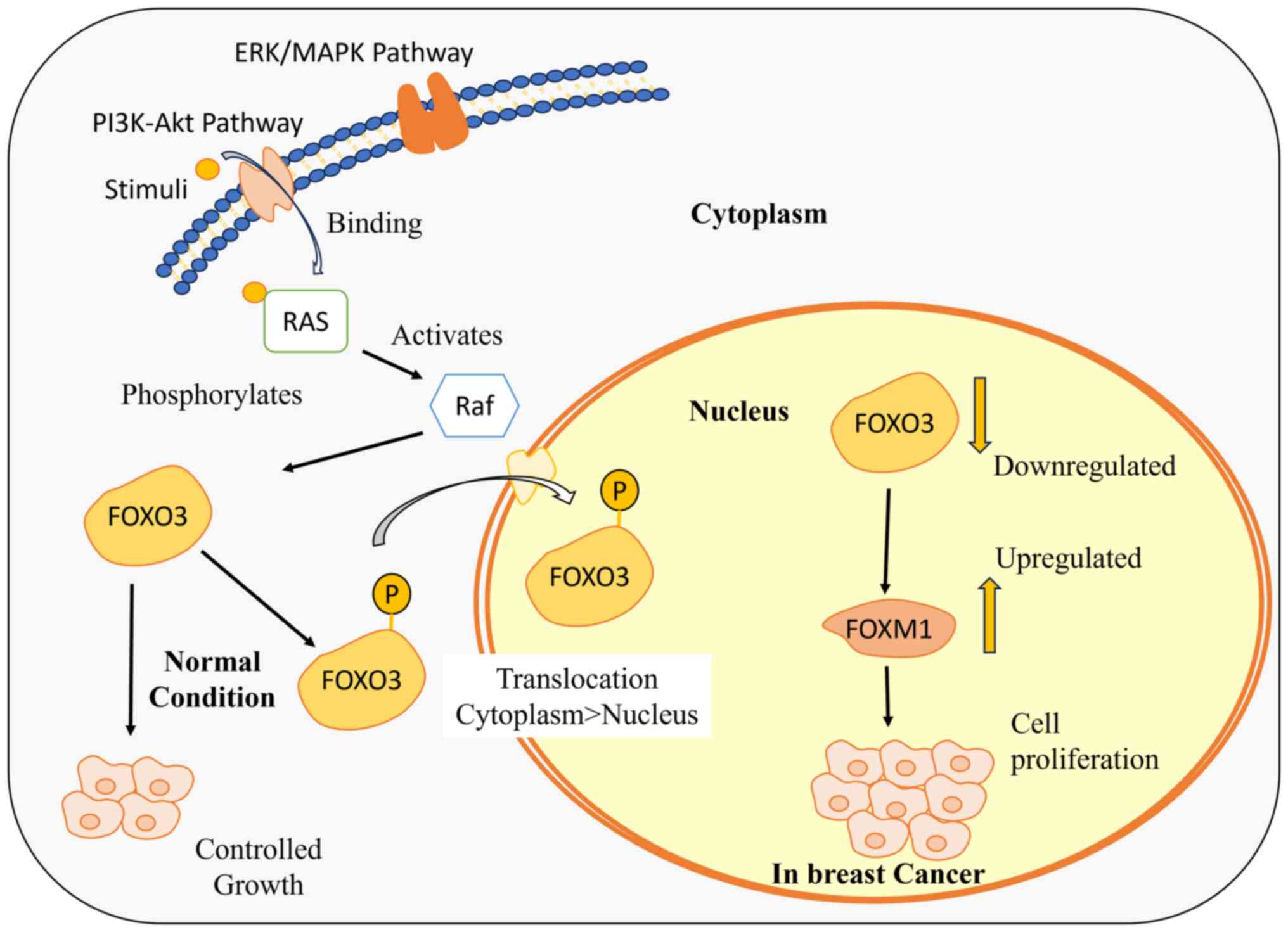
Reduced FOXO protein expression significantly influences breast cancer cell proliferation (31), subject to intricate regulation through processes such as phosphorylation, acetylation and ubiquitination (32). The connection between FOXO and the PI3K/Akt pathway, a critical cell cycle regulator, highlights its function in regulating cellular proliferation and growth (33). The activation of insulin triggers a cascade that involves the production of phosphatidylinositol-3,4,5-triphosphate (PIP3) via PI3K, ultimately leading to phosphorylation of FOXO by Akt (34). This inhibits their DNA-binding domains, thereby reducing the transcriptional activity (31,35). Phosphorylated FOXOs are associated with 14-3-3 proteins, causing nuclear exclusion. Insulin absence restricts FOXO access to the nucleus, impairing its tumor-suppressive activity. The intricate interplay underscores the significance of FOXO proteins in breast cancer regulation, proposing potential therapeutic targets within the PI3K/Akt pathway (36). Various kinases, such as PAK1 and the TAK1-Nemo-like kinase pathway, further modulate FOXO, either inhibiting or promoting its activity (37,38). Understanding these molecular mechanisms provides insights for developing strategies to harness FOXO proteins for controlling breast cancer progression (Fig. 2) (39).
Moreover, FOXO activity is negatively regulated by silent information regulators (SIRTs), which are members of the class III histone deacetylases. SIRTs are acknowledged for their crucial role in cell survival. SIRTs function by downregulating the FOXO levels, thereby controlling the process of apoptosis (40). Oxidative stress results in the nuclear localization of FOXOs. A complex formation occurs in the nucleus by SIRT1, which then interacts with FOXO and deacetylates it, thereby downregulating the tumor-suppressive role FOXO.
The expression of FOXO3a is modulated by the methylation status of its promoter region (41). Liu et al (42) demonstrated that hypermethylation of the FOXO3a promoter by DNMT1 is associated with reduced FOXO3a expression in breast cancer (42). It was further illustrated that FOXO3a suppresses breast cancer stem cell (BCSC) properties and tumorigenicity by inhibiting the FOXM1/SOX2 signaling pathway. Additionally, their findings suggest that SOX2 negatively regulates FOXO3a expression through direct activation of DNMT1, creating a feedback loop. Moreover, blocking DNMT activity suppresses tumor growth by altering the signaling between FOXO3a, FOXM1, and SOX2 proteins in breast cancer. This finding underscores the critical role of the DNMT1/FOXO3a/FOXM1/SOX2 signaling pathway in regulating BCSC properties. It also suggests a promising therapeutic strategy for targeting BCSCs and overcoming drug resistance.
FOXO has emerged as a promising therapeutic target for breast cancer, with recent years witnessing substantial advancements in understanding its role in the disease. Clearer understanding of the processes related to FOXOs would help researchers manipulate and develop more targeted therapies.
FOXM family and breast cancer
FOX proteins exhibit similarity in their DNA-binding domain, characterized by three sheets, three helices, two wings or loops that form a helix-turn-helix motif structure. The second and third helices display structural variations, while the helices and sheets contain conserved regions. The third helix primarily handles DNA binding, with the second wing attaching to the major and minor grooves of DNA (43). Another crucial member of the FOX family, FOXM1, is situated on the 12p13.33 chromosomal band and comprises 10 exons (44). FOXM1 consists of a forkhead domain, an activation domain associated with the C terminus, and a repressor domain linked to the N terminus. The differential splicing of FOXM1 exons gives rise to various variants, including FOXM1A, FOXM1B, FOXM1C and FOXM1D (45). FOXM1 binds to core consensus sequences ‘(A/C) AAACAAC’, and in certain instances, it necessitates interaction with DNA binding factors for transcriptional activity (46). The FOXM1 protein plays a role in diverse processes, encompassing cell proliferation, DNA damage repair and apoptosis. FOXM1 is upregulated in various cancers, highlighting its role as the principal regulator of cell cycle progression, governing various transition points within the cell cycle (47). Additionally, FOXM1 plays a role in regulating gene transcription associated with the cell cycle and regulates various mitotic regulators, including CDC25B and CENPF.
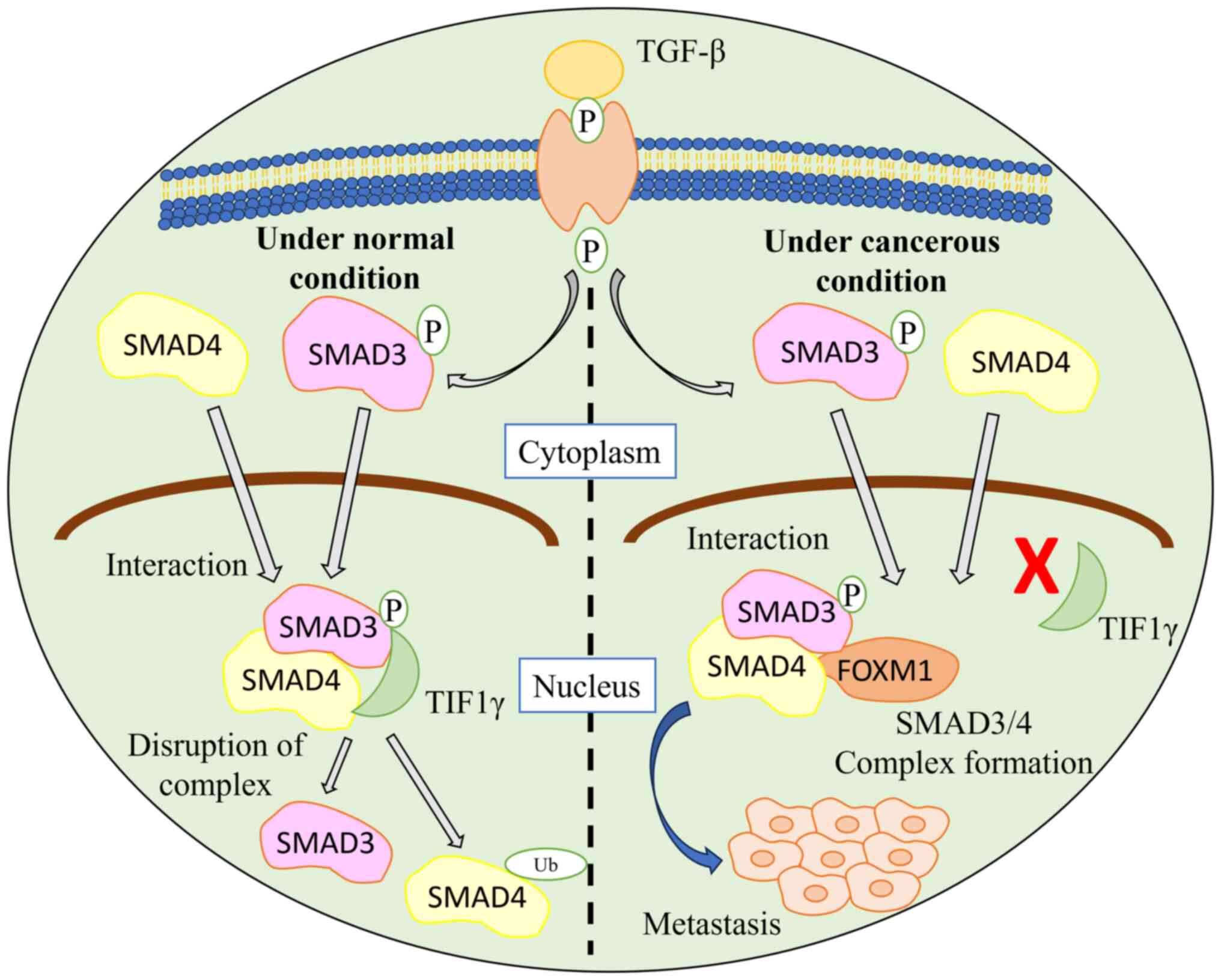
FOXM1 interacts with SMAD3, which activates the TGF-β pathway, resulting in breast cancer cell metastasis. Under normal conditions, the ubiquitin-protein ligase transcriptional intermediary factor 1γ (TIF1γ) interacts with SMAD3 and SMAD4 and causes ubiquitination. This interaction leads to a disruption of the SMAD3/SMAD4 complex. When FOXM1 interacts with SMAD3, the TGF-β pathway is activated as the interaction of SMAD3 and SMAD4 with TIF1 is inhibited, thereby stabilizing the SMAD3/SMAD4 complex (48). The FOXM1/SMAD3 interaction results in breast cancer cell invasion induced by the TGF-β pathway (Fig. 3).
In humans, two types of ER receptors, ERα and Erβ, are responsible for mediating the effect of ER. Although ER binds to both receptors with equal affinity, both exhibit different activities; namely, ERα promote cell proliferation, whereas Erβ has apoptotic and antiproliferative properties (49). FOXM1 has the ability to regulate the levels of ER at both mRNA and protein levels, thereby positively regulating the ER levels by interacting with forkhead-responsive elements situated on the ERα promoter region, thereby influencing its overall regulation.
Human epidermal growth receptor 2 (HER2), a member of the epidermal growth factor receptor family, functions as a tyrosine kinase receptor (50). HER2 is directly correlated with FOXM1 activity. HER2 overexpression in breast cancer results in increased promoter activity of FOXM1, whereas its downregulation upon administration of the HER2 inhibitor (lapatinib) results in a reduced activity of the promoter region of FOXM1 (51).
FOXM1 is an ideal therapeutic target because of its significance in metastasis and disease progression. It is also responsible for relaying resistance to anticancer drugs. However, to date, FOXM1 therapy has not been implemented because of the absence of a comprehensive analysis of the FOXM family. A thorough understanding of FOXM1 mechanisms would help researchers develop biomarkers and therapeutic strategies for early detection and improved treatment strategies.
FOXC family and breast cancer
FOXC proteins are another category of forkhead proteins with distinct DNA binding forkhead domains. Its two members, FOXC1 and FOXC2, play roles in cardiac, embryonic and ocular development (52). In cancer, both members play a significant role in metastasis, cell growth, invasion and angiogenesis (4). FOXC1 is a cell growth regulator in breast cancer and its downregulation results in reduced cell growth, whereas FOXC2 is also involved in carcinogenesis because its increased expression is associated with breast cancer development. FOXC1 is characterized as a single-exon gene situated on the chromosomal band 6p25, encoding a 533 amino acid protein. This encoded protein, localized in the nucleus, engages with DNA and regulates targeted gene expression. Its DNA binding domain contains a ‘winged-helix’ structure. The winged helix's third alpha helix interacts with the major domain at 5′-GTAAATAAA-3′ (53).
Basal-like breast cancer (BLBC) falls under the category of triple-negative breast cancer (TNBC), wherein all three receptors, namely, HER2, progesterone receptor and ER, are not expressed (54). The overexpression of FOXC1 is observed in BLBC, leading to metastasis and invasion (55). A previous study indicated the function of FOXC1 in activating NF-κB signaling. In BLBC, FOXC1 upregulates peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 expression, increasing p65/RelA protein stability (56). This activates the NF-κB signaling pathway which mediates FOXC1 to promote proliferation, invasion and metastasis (55).
In addition to activating the NF-κB signaling pathway, FOXC1 is acknowledged for its role in inducing epithelial-to-mesenchymal transition (EMT) in cancer cells. EMT is a biological process characterized by the loss of cell polarity and intercellular adhesion in embryonic cells, facilitating their migration to various sites for tissue and organ development (57). Overexpression of FOXC1 in MCF12A mammary epithelial cells triggered EMT, as shown by reduced E-cadherin levels. This EMT induction promoted tumor proliferation and migration in BLBC (58,59). Furthermore, EMT is regulated by various transcription factors, including Goosecoid, Snail and Twist, which promote FOXC2 expression in cells undergoing EMT (60). FOXC2 overexpression induces mesenchymal differentiation along with MMP2 and MMP9 expressions. Additionally, FOXC2 is responsible for E-cadherin downregulation, whereas p120-catenin is directly downregulated by FOXC2. p120 is a regulatory protein responsible for stabilizing E-cadherin (61).
The FOXC family is another important category known to regulate tumor progression and metastasis. Relevant research is being conducted, and there is markedly more to discover. Further advances in the interlinkage between FOXC and breast cancer would open the door to advanced therapeutics and treatment strategies.
FOXP family and breast cancer
Similar to other FOX families, the FOXP family is defined by a winged helix domain comprising α-helices, β-sheets and loops or wings, forming a winged-helix forkhead DNA-binding domain (FHD) and a helix-turn-helix motif (62). The FOXP family has four members: FOXP1, FOXP2, FOXP3 and FOXP4, with FOXP1 and FOXP2 having a C-terminal binding domain and FOXP3 and FOXP4 having an N-terminal binding domain (63). Furthermore, all four members exhibit a dimerized C-terminal winged helix FHD, while the FOXP3 dimer is recognized for its stability compared with the others (64).
Depending on the tumor type, FOXP1 acts as an oncogene or tumor suppressor. Similar to FOXA1, the FOXP1 levels are elevated, suggesting its association with ER (65). Elevated FOXP1 levels in MCF7 resulted in increased proliferation along with overexpression of the ER-mediated gene, whereas cells in which the level of FOXP1 was depleted presented reduced growth, with both situations presenting a pro-tumorigenic role in breast cancer (65).
FOXP1, a crucial transcription factor, is intricately linked to the PI3K/Akt signaling pathway which is a key regulator of cellular functions including proliferation and metastasis (66). Akt, a downstream Ser/Thr kinase within this pathway, activates key targets, including p70S6K, which subsequently coregulates diverse cellular functions (67). Notably, FOXP1 levels are modulated by the PI3K/Akt pathway, as proposed by Banham et al (67), who revealed that the absence of Akt significantly reduces FOXP1 levels, whereas the addition of Akt elevates them. Akt does not phosphorylate FOXP1 because of the absence of phosphorylation sites, emphasizing the regulatory role of p70S6K (67) (Fig. 2). A comprehensive understanding of FOXP1 regulatory pathways holds promise for developing innovative therapeutics and treatment strategies.
Liu et al (68) identified the interaction between FOXP2 and FOXA2 and they confirmed that this interaction acts as an EMT suppressor during metastasis in breast cancer cells, highlighting its role as EMT regulator. To understand the in vivo functioning of FOXP2 on the EMT of breast cancer cells, they generated a metastasis model in nude mice and injected the small hairpin (sh)FOXP2 and sh-control lentivirus-infected MCF-7 cells in the tail vein. The results revealed that FOXP2 knockdown promoted metastasis, likely due to the downregulation of E-cadherin and PHF2, both established EMT suppressors indicating that in vivo FOXP2 inhibition could increase the metastasis of breast cancer cells (68).
Conversely, FOXP3 functions as a tumor suppressor by regulating the expression of numerous tumor genes (69). Originally recognized as a molecular marker for regulatory T cells, the involvement of FOXP3 in breast cancer is currently closely associated with metastasis-associated 1 (MTA1), a pivotal protein in downstream gene transcription implicated in breast cancer (70). Functioning as a negative regulator of MTA1 FOXP3 binds to its promoter region, thereby inhibiting its expression and reducing the metastatic capability of the cell (70,71). A thorough investigation into the interplay between FOXP3 and MTA1 could help develop techniques to address metastatic breast cancer and open new avenues for therapeutic advancements in oncology.
The effect of the dysregulated activity of the various FOX proteins and the associated targeted gene are summarized in Table I. While the in vitro and ex vivo studies have helped researchers to comprehend the dysregulated expression of FOX proteins in breast cancer, the current understanding remains incomplete. Therefore, further studies are required to understand the expression levels of FOX proteins utilizing in vivo studies as this approach will be crucial in developing effective therapeutic strategies.
Table I.The effect of the dysregulated activity of the various FOX proteins and the associated targeted gene. |
Overview of miRNA
MiRNAs are small, non-coding RNAs that exert regulatory control over various biological processes. They have a length of 22 nucleotides and function by negatively regulating the mRNA transcription. miRNAs regulate mRNAs by interacting with the 3′ untranslated region (UTR) of the targeted mRNA (76). This interaction suppresses mRNA expression. Apart from the 3′ UTR, miRNA also interacts with other regions such as 5′ UTR, gene promoters and coding sequences (77).
The biogenesis of miRNA involves a two-step process, wherein DNA sequences are transcribed into primary miRNA (pri-miRNA), subsequently processed into precursor and mature miRNAs. The initial step entails transcription of the DNA sequence into pri-miRNA within the nucleus (78). A microprocessor complex, comprising DiGeorge syndrome Critical Region 8 (DGCR8) functioning as an RNA binding domain and Drosha, a ribonuclease III enzyme, is responsible for converting pri-miRNA into pre-miRNA. DGCR8 protein specifically binds to the N6-methylated GGAC sequence in pri-miRNA. This complex then recruits Drosha nuclease, which cleaves the pri-miRNA duplex, resulting in the generation of a pre-miRNA with a characteristic hairpin structure (79,80). Following formation, the pre-miRNA is transported to the cytoplasm via the Exportin 5 (XPO5)/RanGTP complex (81). In the cytoplasm, Dicer, an RNase III enzyme, matures the pre-miRNA by cleaving off the terminal loop, resulting in the formation of mature miRNA (82). The mature miRNA is loaded into the Argonaute protein, which subsequently regulates the targeted mRNA.
miRNAs play regulatory roles in various processes, including cell proliferation and apoptosis. The involvement of miRNAs in breast cancer, either as tumor suppressor miRNAs (tsmiRs) or oncogenic miRNAs (oncomiRs), has been underscored in numerous studies. OncomiRs are miRNAs capable of downregulating the expression of tumor suppressors and are therefore elevated in breast cancer (83). tsmiRs are miRNAs capable of downregulating the expression of oncogenes and are thereby downregulated in breast cancer (84). miRNAs are known for their regulatory roles in cellular mechanisms, including cell proliferation, metastasis, invasion and apoptosis; therefore, the altered miRNAs levels can induce cancer-like conditions. For instance, miR-16 and miR-497 are known to inhibit cell proliferation, thereby acting as tsmiRs (85). Contrarily, miR-200c/141 works as an oncogene, thereby promoting the metastasis of breast cancer cells (86).
Interlinkage of miRNA and FOX proteins in breast cancer
FOX proteins are a group of transcription factors that play a regulatory role in controlling gene expression. FOX proteins function as oncogenes and tumor suppressors; therefore, any dysregulation in FOX protein levels results in cancer-like conditions.
On the other hand, miRNA is a small non-coding RNA that works by regulating target mRNA expression. In breast cancer, miRNAs are known to function as either oncomiRs or tsmiRs, thereby promoting the pathological condition. miRNA overexpression or downregulation results in increased cell proliferation and metastasis, resulting in tumorigenesis.
Research is being conducted to understand the interlinkage between miRNA and various classes of FOX proteins as miRNAs regulate the expression of FOX proteins, which leads to breast cancer initiation and progression. Therefore, detailed insights into the crosslinks between miRNA and FOX proteins and the role of miRNA in modulating the activity of FOX proteins would help in advancing the effective treatment for breast cancer.
miR-132, a tumor suppressor, is downregulated in breast cancer. It regulates cell growth, proliferation and apoptosis, and is known to downregulate the expression of FOXA1 (87). The overexpression of miR-132 downregulates FOXA1, thereby affecting the expression of LIPG (subsequent target of FOXA1). miR-132 downregulation results in increased cell proliferation along with elevated levels of FOXA1 and LIPG, thereby resulting in tumorigenesis (88). In a study conducted by Wang et al (88), it was observed that introducing miR-132 significantly hindered colony formation in MDA-MB-468 cells compared with the control group whereas suppressing miR-132 enhanced colony formation efficiency in these cells. Reducing FOXA1 expression led to a notable decrease in colony formation efficiency in breast cancer cells compared with the anti-miR-132 group. The aforementioned study revealed an inverse correlation between miR-132 and FOXA1 expression in breast cancer tissues. Furthermore, miR-132 was shown to suppress FOXA1 expression in breast cancer cells. These findings suggested that miR-132 downregulation might contribute to FOXA1 upregulation in breast cancer. Therefore, targeting miR-132 and FOXA1 together could hold promise as therapeutic strategy for breast cancer treatment. miR-802 is a tumor suppressor miRNA, responsible for regulating cell proliferation (89). FOXM1, an oncogene, is negatively regulated by miR-802 because it contains miR-802 binding sites. miR-802 interacts with the 3′ UTR of FOXM1, downregulating its expression. The upregulation of miR-802 results in the reduced expression of FOXM1, thereby controlling cell proliferation, whereas the downregulated expression of miR-802 elevates FOXM1 expression, contributing to cancer-like conditions. Yuan and Wang (90) delved into the expression pattern of miR-802 in cancerous vs. adjacent non-cancerous tissues, revealing a significant decrease in miR-802 levels in cancer tissues. It was demonstrated that increasing miR-802 levels significantly reduced the ability of MCF-7 cells to proliferate after transfection, compared with control cells. To confirm that miR-802 directly targets FoxM1, they created a luciferase reporter vector containing potential miR-802 binding sites within the 3′ UTR of the FoxM1 gene. Their outcomes showed that increasing miR-802 levels significantly reduced luciferase activity in MCF-7 cells when the reporter contained the FoxM1 3′ UTR. These findings provide initial evidence that boosting miR-802 expression hinders breast cancer cell proliferation both in cell culture (in vitro) and in animal models (in vivo), likely by regulating FoxM1 expression. This highlights the significance of miR-802 in breast cancer development and suggests its potential as a target for breast cancer therapy. miR-199a, identified as an oncogenic miRNA, exhibits elevated levels in breast cancer cells. The tumor suppressor FOXP2 is negatively regulated by miR-199a (91). Overexpression of miR-199a causes a decrease in FOXP2 protein expression in breast cancer cells, consequently promoting increased proliferation, survival and metastasis of breast cancer cells. Cuiffo et al (92) demonstrated that manipulating specific miRNAs or silencing FOXP2 resulted in increased BCSC properties, leading to enhanced tumor initiation and metastasis. They validated their initial findings on FOXP2 expression using independent reverse transcription-quantitative PCR (ΔΔCq) analysis, revealing a significant decrease in FOXP2 protein levels in BCC199a, BCC199a/214 and BCCMSC cell lines. Since the decrease in FOXP2 was associated with maintaining progenitor cell identity and inhibiting differentiation in these cellular contexts, it was suggested that downregulation of FOXP2 might contribute to the emergence of CSC-like phenotypes in BCC199a, BCC199a/214 and BCCMSC. These findings contribute to a holistic comprehension of miRNA-mediated regulation in breast cancer, as outlined in Table II.
FOX proteins and drug resistance
Despite the condition's heterogeneity, various therapeutics are employed to treat breast cancer. Although beneficial, the major difficulty faced by researchers is drug resistance. After completing a few drug cycles, patients start to develop resistance against the administered drug, thereby reducing its efficiency. The development of resistance involves various signaling pathways; recently, FOX proteins are also observed in this light. Alterations in the expression levels of various FOX proteins contribute to the development of resistance against anticancer drugs. Endocrine therapeutics are administered to target and regulate the estrogen response in breast cancer cells. Although favorable, the major setback of endocrine therapies revolves around the emergence of resistance against anticancer drugs. Tumors that develop resistance can regulate the elevated ER expression by employing various processes, including drug efflux, modulation of ER cofactor expression, and activation of other pathways, including growth factor signaling (97).
FOXA1 exhibits a positive association with all ER+ luminal breast cancer cells and works by interacting with DNA to open up chromatin, which then elevates the ER interaction to make its response element more accessible. The overall increased interaction is responsible for mediating resistance to endocrine therapeutics, including tamoxifen (98).
Tamoxifen is an FDA-approved ER modulator used as an endocrine therapy for breast cancer treatment. Tamoxifen does not entirely inhibit the function of ER; instead, it binds to ER, and this complex is recruited to chromatin, subsequently leading to the suppression of ER target gene transcription. Similar to normal estrogen-bound ER, tamoxifen-bound ER also requires FOXM1 to access the chromatin and perform its function (99).
Dysregulated FOXA1 expression promotes resistance to tamoxifen, thereby limiting its effectiveness. The overexpression of FOXA1 leads to the upregulation of IL-8, a cytokine responsible for metastases and survival of cancer cells and contributes to resistance to ER inhibitors. Fu et al (98) investigated the contribution of FOXA1 to endocrine therapy resistance in ER-positive (ER+) breast cancer. They employed various endocrine-resistant (Endo-R) breast cancer cell lines derived from parental ER+ lines. Notably, they observed elevated FOXA1 expression in these Endo-R cell lines compared with their ER+ counterparts. Additionally, elevated levels of FOXA1 mRNA were predictive of unfavorable outcomes in patients with ER+ tumor receiving tamoxifen. Importantly, IL-8 emerged as a critical mediator of the FOXA1/ER transcriptional reprogramming process, promoting the growth and invasion of Endo-R cells. The study suggests that targeting the IL-8 signaling pathway might be a promising therapeutic approach for treating ER+ breast cancers with elevated levels of FOXA1.
The downregulation of FOXA1 results in upregulation of IL-6, thus promoting cancer-like properties in tamoxifen-resistant breast cancer cells (100). In a study by Yamaguchi et al (100), tamoxifen-resistant (TAM-R) breast cancer cells were developed by prolonged exposure of ER+ MCF7 cells to tamoxifen. Interestingly, TAM-R cells displayed lower levels of both ERα, a major ER form in breast cancer, and its co-regulator FOXA1. By contrast, these cells displayed increased activation of the transcription factor NF-κB and its target gene, IL6. Through experimental interventions, stable expression of FOXA1, but not ERα, led to reduced IL6 levels in both MDA-MB-231 cells (lacking FOXA1 and ERα) and TAM-R cells, without significantly affecting NF-κB activity. Conversely, depletion of FOXA1 increased IL6 expression in MCF7 cells.
FOXM1, a key regulator in therapeutic resistance among breast cancer cells, is associated with various cancer conditions due to its influence on cell cycle regulation, autophagy and senescence. Elevated FOXM1 levels contribute to drug resistance, notably against the anthracycline drug epirubicin (101). OTU domain-containing ubiquitin aldehyde-binding protein 1 (OTUB1) acts as a regulator, with upregulated FOXM1 resulting in increased cell proliferation and resistance development against epirubicin. Lys48-linked polyubiquitin chains, pivotal in proteasomal regulation, interact with FOXM1 to modulate its levels. However, FOXM1-OTUB1 interaction prevails, enhancing FOXM1 expression and conferring resistance (102). Elevated FOXM1 levels also facilitate improved DNA damage repair induced by epirubicin. Additionally, FOXM1 regulates antiapoptotic genes XIAP and survivin, fostering resistance against doxorubicin, epirubicin, docetaxel and paclitaxel (103). Understanding the intricate role of FOXM1 provides insights for targeted therapeutic interventions.
The FOXO3-FOXM1 axis serves as an informative marker to comprehend the resistance of drug, as its dysregulation impacts the cellular fate. FOXO3 is recognized as a tumor suppressor that regulates cell growth, proliferation and apoptosis (30). The activity of FOXO3 is downregulated by Ras-Raf-MEK-extracellular signal-regulated kinase (ERK) and phosphatidylinositol 3-kinase (PI3K)-Akt/Protein Kinase B (PKB) signaling pathways in response to various external stimuli and growth factors. Apart from all the targets, FOXO3 downregulates the FOXM1 expression levels and competes with FOXM1 to interact with the same binding sites at the gene promoters (102). FOXM1 is an oncogene responsible for cell growth and proliferation, thereby promoting tumorigenesis (104). The downregulation of FOXO3 induces an upregulation of FOXM1, promoting drug efflux and cell survival. This upregulation contributes to an increase in reactive oxygen species levels, ultimately leading to the development of resistance against the administered drug (105) (Fig. 4).
FOX therapeutics: Current trends and future perspectives
Chemotherapy, a widely used treatment for breast cancer, targets and destroys cancer cells, with ongoing research aimed at enhancing its effectiveness against the disease. Distinct subclasses within the FOX family are identified as either oncogenes or tumor suppressors, exerting regulatory control over cellular fate through diverse mechanisms. This underscores their pivotal role in the initiation and progression of tumors (106). Targeting FOX proteins as a therapeutic intervention holds promise in the development of drugs responsible for regulation of FOX protein levels, thereby effectively inhibiting the growth of breast cancer cells which may potentially improve the efficacy of existing treatment strategies.
FOXM1 emerges as a crucial therapeutic target for breast cancer, particularly in HER2-positive cases. Lapatinib, an FDA-approved tyrosine kinase inhibitor, effectively downregulates elevated HER2 levels, concurrently decreasing FOXM1 expression. Francis et al (51) elucidated the connection between HER2 and FOXM1, establishing the ability of lapatinib to regulate FOXM1 at both mRNA and protein levels. Alternatively, honokiol, a plant-derived compound, demonstrates anti-inflammatory, antioxidative and anticancer properties, including FOXM1 inhibition. Honokiol disrupts the positive feedback loop regulating FOXM1 transcription, leading to reduced mRNA and protein levels. This dual approach, targeting FOXM1 with lapatinib and honokiol, offers a promising avenue for developing comprehensive breast cancer therapeutics, complementing existing strategies by Halasi et al (107).
Imipramine blue (IB), an anticancer derivative of the FDA-approved antidepressant imipramine, demonstrates efficacy in downregulating elevated FOXM1 expression in breast cancer, as demonstrated by Rajamanickam et al (108). The ability of IB to target FOXM1 and inhibit homologous recombination-mediated DNA repair positions it as a potential therapeutic agent for regulating tumor growth and metastasis in breast cancer. Integrating IB into treatment protocols could enable effective targeting of FOXM1 and improve the outcomes of breast cancer therapies.
Combination therapy has become pivotal in cancer treatment due to the limited efficacy of single-agent targeted drugs in clinical trials despite the expanding array of options available (109). Combining targeted therapies with hormone therapies (endocrine treatments) has been successful in improving the outlook for patients with breast cancer. However, some combinations of these drugs may have a higher chance of causing adverse effects. Therefore, identifying precise combinations and dosing regimens to optimize therapeutic efficacy while minimizing side effects is crucial in both preclinical and clinical investigations (110). A notable success in breast cancer combination therapy involves incorporating the anti-HER2 monoclonal antibody trastuzumab into first-line chemotherapy, significantly enhancing treatment benefits in patients with HER2-overexpressing metastatic breast cancer (111). Currently, the trastuzumab and paclitaxel combination is a potent regimen for treating low-risk HER2-positive tumors, yielding sustained positive outcomes (112). In a recent study, researchers investigated the synergistic potential of FOXM1 inhibitors alongside proteasome inhibitors such as Bortezomib, revealing significant inhibition of proliferation in both ER+ and TNBC (113). Similarly, the combined efficacy of FOXM1 inhibitors with CDK4/6 inhibitors including palbociclib, ribociclib and abemaciclib was demonstrated in ER+ breast cancer cells (113). These findings offer promise for advancing translational efforts and initiating clinical trials investigating agents targeting FOXM1, potentially improving outcomes for patients with breast cancer.
Targeting Fox proteins, transcription factors linked to breast cancer, is a challenge due to selectivity issues (4). Ideally, therapies should target only the cancer-promoting functions of Fox proteins in tumors, leaving out the healthy tissues. However, this objective presents several key challenges. Inhibitor off-target effects can disrupt similar proteins in healthy cells, causing side effects. Lin et al (114) demonstrated that FOXC1 promotes EMT and metastasis in TNBC cell lines. Their study revealed that inhibiting FOXC1 with a specific inhibitor significantly suppressed TNBC cell invasion and metastasis in preclinical models. However, a critical challenge identified in the aforementioned study is the potential for off-target effects of the inhibitor. Further optimization is necessary to minimize adverse consequences on healthy cells (114). FOXA1 plays a crucial role in ER signaling and tumor progression in hormone receptor (HR)+ breast cancer. Robinson et al (17) employed whole-genome analysis of HR+ breast cancer and identified FOXA1 as a key regulator of ER-mediated transcription. While targeting FOXA1 holds promise for disrupting ER signaling and inhibiting tumor growth, a major challenge lies in developing selective inhibitors that specifically target FOXA1 within cancer cells without compromising its normal functions in ER+ healthy breast tissue (17). It is important to understand that the functional similarity between FOX proteins in healthy and cancerous cells possess a significant challenge. The development of inhibitors that precisely target the tumorigenic functions of FOX proteins is challenging. This underscores the challenge of specifically targeting the cancer-promoting functions of FOX proteins without affecting their essential physiological roles in healthy tissues; therefore, a deeper understanding of Fox protein activity unique to cancer cells is crucial to overcome this (4).
Although therapeutics targeting specific miRNAs have not yet been applied clinically in breast cancer, notable progress is being made. For example, a miR-122 antagonist is currently undergoing Phase II clinical trials for hepatitis C treatment, suggesting the potential for miRNAs to serve as viable therapeutic targets in breast cancer (115). Efforts are also being directed towards developing therapeutics targeting miR-21 and miR-221. These microRNAs have been linked to drug resistance in breast cancer, and scientists are investigating their potential as therapeutic targets for hepatocellular carcinoma and other cancers (116). Clinical trials testing miRNA inhibitors as a treatment for patients with breast cancer with multidrug resistance could be initiated in the near future. Addressing the epigenetic alterations associated with drug resistance in breast cancer presents significant challenges. However, histone deacetylation inhibitors are emerging as promising candidates for all breast cancer subtypes (117,118). It is crucial to proceed cautiously, as these inhibitors may inadvertently alter the expression of key receptors such as ERα, HER2 and MDR1, potentially exacerbating drug resistance. Ongoing clinical trials of various histone deacetylation inhibitors will provide valuable insights into their efficacy and safety profiles (118,119).
Understanding the factors involved in cancerous cell initiation, invasion and metastasis is essential for developing new treatment strategies and improving existing ones. Numerous factors have been discovered so far that have been targeted to develop various therapeutics. FOX proteins are an ideal biomarker and therapeutic target that would aid in understanding and improving the existing therapies for breast cancer. Clinical trials targeting FOX proteins, therapeutics and breast cancer, that are currently underway are summarized in Table III.
Conclusions
In recent years, significant advancements have been achieved in the realm of breast cancer research, owing to researchers' adeptness in selectively addressing multiple signaling pathways whose dysregulation contributes to the initiation and progression of breast cancer. Apart from the pathways, diverse transcription factors are involved in breast cancer tumorigenesis. FOX proteins function as transcription factors and have been linked to numerous processes, including cell growth and apoptosis. The altered levels of distinct subclasses of FOX proteins can act as either oncogenes or tumor suppressors, thus inducing abnormal proliferation in breast cancer cells. miRNAs regulate FOX proteins, thereby controlling their expression, and the dysregulation of FOX protein levels caused by altered miRNA levels contributes to tumorigenesis.
Research focusing on FOX factors has provided valuable insights into the underlying mechanisms driving cancer progression. FOX factors often play diverse roles in cancer progression, influencing various aspects of tumor development and metastasis. Understanding the specific functions of FOX factors in cancer modulation and identifying potential therapeutic targets capable of modulating their activities could expand treatment options for cancer patients. Inhibiting FOX factors or the pathways they regulate holds potential for reversing drug resistance, overcoming immune evasion and halting metastatic spread. Therapeutic strategies targeting FOX factors may involve inhibitors of the factors themselves, the kinases involved in FOX-mediated pathways, or agents utilizing RNA interference to silence FOX factor expression. These approaches represent promising avenues for treating various types of cancer and offer hope for improved patient outcomes. Additionally, FOX proteins can potentially relay drug resistance, making them an appropriate therapeutic target, with only few FOX proteins being investigated as potential therapeutic targets. Thus, a thorough understanding of the mechanisms behind the various classes of FOX proteins would help to develop drugs that target the FOX proteins, control their levels, and thereby assist in the development of efficient treatment approaches.
Acknowledgements
Not applicable.
Funding
This research has been funded by Scientific Research Deanship at University of Ha'il-Saudi Arabia through project number (RG-23 131).
Availability of data and materials
Not applicable.
Authors' contributions
SA and MAK wrote the major parts of the manuscript and prepared the figures and tables. MZ and MAH revised the manuscript. NI, SA, AA and SK performed the bibliographic research and prepared the table and figures. S, SK and MZN conceptualized the study and oversaw the process. All authors contributed to writing the manuscript. All authors read and approved the final manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Weiderpass E and Stewart BW: World cancer report: Cancer research for cancer prevention. International Agency for Research on Cancer; Lyon: 2020 | |
|
Feng Y, Spezia M, Huang S, Yuan C, Zeng Z, Zhang L, Ji X, Liu W, Huang B, Luo W, et al: Breast cancer development and progression: risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 5:77–106. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Laissue P: The forkhead-box family of transcription factors: Key molecular players in colorectal cancer pathogenesis. Mol Cancer. 18:52019. View Article : Google Scholar : PubMed/NCBI | |
|
Bach DH, Long NP, Luu TT, Anh NH, Kwon SW and Lee SK: The dominant role of forkhead box proteins in cancer. Int J Mol Sci. 19:32792018. View Article : Google Scholar : PubMed/NCBI | |
|
Myatt SS and Lam EW: The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Weigel D, Jürgens G, Küttner F, Seifert E and Jäckle H: The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 57:645–658. 1989. View Article : Google Scholar : PubMed/NCBI | |
|
Vaidya HJ, Briones Leon A and Blackburn CC: FOXN1 in thymus organogenesis and development. Eur J Immunol. 46:1826–1837. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Lam EW and Gomes AR: Forkhead box transcription factors in cancer initiation, progression and chemotherapeutic drug response. Front Oncol. 4:3052014. View Article : Google Scholar : PubMed/NCBI | |
|
Li C, Zhang K, Chen J, Chen L, Wang R and Chu X: MicroRNAs as regulators and mediators of forkhead box transcription factors function in human cancers. Oncotarget. 8:12433–12450. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Seachrist DD, Anstine LJ and Keri RA: FOXA1: A pioneer of nuclear receptor action in breast cancer. Cancers (Basel). 13:52052021. View Article : Google Scholar : PubMed/NCBI | |
|
Czerny CC, Borschel A, Cai M, Otto M and Hoyer-Fender S: FOXA1 is a transcriptional activator of Odf2/Cenexin and regulates primary ciliation. Sci Rep. 12:214682022. View Article : Google Scholar : PubMed/NCBI | |
|
Cirillo LA and Zaret KS: Specific interactions of the wing domains of FOXA1 transcription factor with DNA. J Mol Biol. 366:720–724. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Bernardo GM and Keri RA: FOXA1: A transcription factor with parallel functions in development and cancer. Biosci Rep. 32:113–130. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Dai X, Cheng H, Bai Z and Li J: Breast cancer cell line classification and its relevance with breast tumor subtyping. J Cancer. 8:3131–3141. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Liu Y, Zhao Y, Skerry B, Wang X, Colin-Cassin C, Radisky DC, Kaestner KH and Li Z: Foxa1 is essential for mammary duct formation. Genesis. 54:277–285. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Brisken C and O'Malley B: Hormone action in the mammary gland. Cold Spring Harb Perspect Biol. 2:a0031782010. View Article : Google Scholar : PubMed/NCBI | |
|
Robinson JL, Macarthur S, Ross-Innes CS, Tilley WD, Neal DE, Mills IG and Carroll JS: Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J. 30:3019–3027. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Yang YA, Zhao JC, Fong KW, Kim J, Li S, Song C, Song B, Zheng B, He C and Yu J: FOXA1 potentiates lineage-specific enhancer activation through modulating TET1 expression and function. Nucleic Acids Res. 44:8153–8164. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Bernardo GM, Lozada KL, Miedler JD, Harburg G, Hewitt SC, Mosley JD, Godwin AK, Korach KS, Visvader JE, Kaestner KH, et al: FOXA1 is an essential determinant of ERalpha expression and mammary ductal morphogenesis. Development. 137:2045–2054. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Takaku M, Grimm SA, De Kumar B, Bennett BD and Wade PA: Cancer-specific mutation of GATA3 disrupts the transcriptional regulatory network governed by Estrogen Receptor alpha, FOXA1 and GATA3. Nucleic Acids Res. 48:4756–4768. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Ghosh S, Gu F, Wang CM, Lin CL, Liu J, Wang H, Ravdin P, Hu Y, Huang TH and Li R: Genome-wide DNA methylation profiling reveals parity-associated hypermethylation of FOXA1. Breast Cancer Res Treat. 147:653–659. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Slebe F, Rojo F, Vinaixa M, García-Rocha M, Testoni G, Guiu M, Planet E, Samino S, Arenas EJ, Beltran A, et al: FoxA and LIPG endothelial lipase control the uptake of extracellular lipids for breast cancer growth. Nat Commun. 7:111992016. View Article : Google Scholar : PubMed/NCBI | |
|
Anzai E, Hirata K, Shibazaki M, Yamada C, Morii M, Honda T and Yamaguchi N and Yamaguchi N: FOXA1 induces E-cadherin expression at the protein level via suppression of slug in epithelial breast cancer cells. Biol Pharm Bull. 40:1483–1489. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Ambrosone CB and Higgins MJ: Relationships between breast feeding and breast cancer subtypes: Lessons learned from studies in humans and in mice. Cancer Res. 80:4871–4877. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Xia K, Huang W, Zhao X, Huang X, Chen Y, Yu L and Tan Y: Increased FOXA1 levels induce apoptosis and inhibit proliferation in FOXA1-low expressing basal breast cancer cells. Am J Cancer Res. 12:2641–2658. 2022.PubMed/NCBI | |
|
Cantor JR and Sabatini DM: Cancer cell metabolism: One hallmark, many faces. Cancer Discov. 2:881–898. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Wang Y, Zhou Y and Graves DT: FOXO transcription factors: Their clinical significance and regulation. Biomed Res Int. 2014:9253502014.PubMed/NCBI | |
|
Jiramongkol Y and Lam EW: FOXO transcription factor family in cancer and metastasis. Cancer Metastasis Rev. 39:681–709. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Dumont SN, Lazar AJ, Bridge JA, Benjamin RS and Trent JC: PAX3/7-FOXO1 fusion status in older rhabdomyosarcoma patient population by fluorescent in situ hybridization. J Cancer Res Clin Oncol. 138:213–220. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Parry P, Wei Y and Evans G: Cloning and characterization of the t(X;11) breakpoint from a leukemic cell line identify a new member of the forkhead gene family. Genes Chromosomes Cancer. 11:79–84. 1994. View Article : Google Scholar : PubMed/NCBI | |
|
Guttilla IK and White BA: Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 284:23204–23216. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Bullock M: FOXO factors and breast cancer: Outfoxing endocrine resistance. Endocr Relat Cancer. 23:R113–R130. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Farhan M, Wang H, Gaur U, Little PJ, Xu J and Zheng W: FOXO signaling pathways as therapeutic targets in cancer. Int J Biol Sci. 13:815–827. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Di Blasio L, Gagliardi PA, Puliafito A and Primo L: Serine/threonine kinase 3-phosphoinositide-dependent protein Kinase-1 (PDK1) as a key regulator of cell migration and cancer dissemination. Cancers (Basel). 9:252017. View Article : Google Scholar : PubMed/NCBI | |
|
Shaw RJ and Cantley LC: Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 441:424–430. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Tzivion G, Dobson M and Ramakrishnan G: FoxO transcription factors; regulation by AKT and 14-3-3 proteins. Biochim Biophys Acta. 1813:1938–1945. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Kim S, Kim Y, Lee J and Chung J: Regulation of FOXO1 by TAK1-Nemo-like kinase pathway. J Biol Chem. 285:8122–8129. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Liu H, Liu K and Dong Z: The role of p21-activated kinases in cancer and beyond: Where are we heading? Front Cell Dev Biol. 9:6413812021. View Article : Google Scholar : PubMed/NCBI | |
|
Khan MA, Massey S, Ahmad I, Sada f, Akhter N, Habib M, Mustafa S, Deo SVS and Husain SA: FOXO1 gene downregulation and promoter methylation exhibits significant correlation with clinical parameters in Indian breast cancer patients. Front Genet. 13:8429432022. View Article : Google Scholar : PubMed/NCBI | |
|
Peck B, Chen CY, Ho KK, Di Fruscia P, Myatt SS, Coombes RC, Fuchter MJ, Hsiao CD and Lam EW: SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol Cancer Ther. 9:844–855. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Gong C, Yao S, Gomes AR, Man EP, Lee HJ, Gong G, Chang S, Kim SB, Fujino K, Kim SW, et al: BRCA1 positively regulates FOXO3 expression by restricting FOXO3 gene methylation and epigenetic silencing through targeting EZH2 in breast cancer. Oncogenesis. 5:e2142016. View Article : Google Scholar : PubMed/NCBI | |
|
Liu H, Song Y, Qiu H, Liu Y, Luo K, Yi Y, Jiang G, Lu M, Zhang Z, Yin J, et al: Downregulation of FOXO3a by DNMT1 promotes breast cancer stem cell properties and tumorigenesis. Cell Death Differ. 27:966–983. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Sanders DA, Gormally MV, Marsico G, Beraldi D, Tannahill D and Balasubramanian S: FOXM1 binds directly to non-consensus sequences in the human genome. Genome Biol. 16:1302015. View Article : Google Scholar : PubMed/NCBI | |
|
Korver W, Roose J, Heinen K, Weghuis DO, de Bruijn D, van Kessel AG and Clevers H: The human TRIDENT/HFH-11/FKHL16 gene: Structure, localization, and promoter characterization. Genomics. 46:435–442. 1997. View Article : Google Scholar : PubMed/NCBI | |
|
Kalathil D, John S and Nair AS: FOXM1 and cancer: Faulty cellular signaling derails homeostasis. Front Oncol. 10:6268362021. View Article : Google Scholar : PubMed/NCBI | |
|
Ye H, Kelly TF, Samadani U, Lim L, Rubio S, Overdier DG, Roebuck KA and Costa RH: Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 17:1626–1641. 1997. View Article : Google Scholar : PubMed/NCBI | |
|
Halasi M and Gartel AL: FOX(M1) news-it is cancer. Mol Cancer Ther. 12:245–254. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Xue J, Lin X, Chiu WT, Chen YH, Yu G, Liu M, Feng XH, Sawaya R, Medema RH, Hung MC and Huang S: Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-β-dependent cancer metastasis. J Clin Invest. 124:564–579. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Speirs V and Walker RA: New perspectives into the biological and clinical relevance of oestrogen receptors in the human breast. J Pathol. 211:499–506. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Iqbal N and Iqbal N: Human epidermal growth factor Receptor 2 (HER2) in cancers: Overexpression and therapeutic implications. Mol Biol Int. 2014:8527482014. View Article : Google Scholar : PubMed/NCBI | |
|
Francis RE, Myatt SS, Krol J, Hartman J, Peck B, McGovern UB, Wang J, Guest SK, Filipovic A, Gojis O, et al: FoxM1 is a downstream target and marker of HER2 overexpression in breast cancer. Int J Oncol. 35:57–68. 2009.PubMed/NCBI | |
|
Chen X, Wei H, Li J, Liang X, Dai S, Jiang L, Guo M, Qu L, Chen Z, Chen L and Chen Y: Structural basis for DNA recognition by FOXC2. Nucleic Acids Res. 47:3752–3764. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Pierrou S, Enerbäck S and Carlsson P: Selection of high-affinity binding sites for sequence-specific, DNA binding proteins from random sequence oligonucleotides. Anal Biochem. 229:99–105. 1995. View Article : Google Scholar : PubMed/NCBI | |
|
Yin L, Duan JJ, Bian XW and Yu SC: Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 22:612020. View Article : Google Scholar : PubMed/NCBI | |
|
Han B, Bhowmick N, Qu Y, Chung S, Giuliano AE and Cui X: FOXC1: An emerging marker and therapeutic target for cancer. Oncogene. 36:3957–3963. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Wang J, Ray PS, Sim MS, Zhou XZ, Lu KP, Lee AV, Lin X, Bagaria SP, Giuliano AE and Cui X: FOXC1 regulates the functions of human basal-like breast cancer cells by activating NF-kappaB signaling. Oncogene. 31:4798–4802. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Nieto MA: Epithelial plasticity: A common theme in embryonic and cancer cells. Science. 342:12348502013. View Article : Google Scholar : PubMed/NCBI | |
|
Bloushtain-Qimron N, Yao J, Snyder EL, Shipitsin M, Campbell LL, Mani SA, Hu M, Chen H, Ustyansky V, Antosiewicz JE, et al: Cell type-specific DNA methylation patterns in the human breast. Proc Natl Acad Sci USA. 105:14076–14081. 2008. View Article : Google Scholar : PubMed/NCBI | |
|
Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA, et al: Single cell profiling of circulating tumor cells: Transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One. 7:e337882012. View Article : Google Scholar : PubMed/NCBI | |
|
Lindley LE and Briegel KJ: Molecular characterization of TGFbeta-induced epithelial-mesenchymal transition in normal finite lifespan human mammary epithelial cells. Biochem Biophys Res Commun. 399:659–664. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
Lamouille S, Xu J and Derynck R: Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 15:178–196. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Clark KL, Halay ED, Lai E and Burley SK: Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 364:412–420. 1993. View Article : Google Scholar : PubMed/NCBI | |
|
Perumal K, Dirr HW and Fanucchi S: A single amino acid in the hinge loop region of the FOXP forkhead domain is significant for dimerisation. Protein J. 34:111–121. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H and Chen L: Structure of the forkhead domain of FOXP2 bound to DNA. Structure. 14:159–166. 2006. View Article : Google Scholar : PubMed/NCBI | |
|
Shigekawa T, Ijichi N, Ikeda K, Horie-Inoue K, Shimizu C, Saji S, Aogi K, Tsuda H, Osaki A, Saeki T and Inoue S: FOXP1, an estrogen-inducible transcription factor, modulates cell proliferation in breast cancer cells and 5-year recurrence-free survival of patients with tamoxifen-treated breast cancer. Horm Cancer. 2:286–297. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Beelen K, Hoefnagel LD, Opdam M, Wesseling J, Sanders J, Vincent AD, van Diest PJ and Linn SC: PI3K/AKT/mTOR pathway activation in primary and corresponding metastatic breast tumors after adjuvant endocrine therapy. Int J Cancer. 135:1257–1263. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Banham AH, Beasley N, Campo E, Fernandez PL, Fidler C, Gatter K, Jones M, Mason DY, Prime JE, Trougouboff P, et al: The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p. Cancer Res. 61:8820–8829. 2001.PubMed/NCBI | |
|
Liu Y, Chen T, Guo M, Li Y, Zhang Q, Tan G, Yu L and Tan Y: FOXA2-interacting FOXP2 prevents epithelial-mesenchymal transition of breast cancer cells by stimulating E-cadherin and PHF2 transcription. Front Oncol. 11:6050252021. View Article : Google Scholar : PubMed/NCBI | |
|
Sada f, Akhter N, Alharbi RA, Sindi AAA, Najm MZ, Alhumaydhi FA, Khan MA, Deo SVS and Husain SA: Epigenetic alteration and its association with downregulated FOXP3 gene in indian breast cancer patients. Front Genet. 12:7814002021. View Article : Google Scholar : PubMed/NCBI | |
|
Liu C, Han J, Li X, Huang T, Gao Y, Wang B, Zhang K, Wang S, Zhang W, Li W, et al: FOXP3 inhibits the metastasis of breast cancer by downregulating the expression of MTA1. Front Oncol. 11:6561902021. View Article : Google Scholar : PubMed/NCBI | |
|
Ma B, Miao W, Xiao J, Chen X, Xu J and Li Y: The role of FOXP3 on tumor metastasis and its interaction with traditional Chinese medicine. Molecules. 27:67062022. View Article : Google Scholar : PubMed/NCBI | |
|
Dai X, Cheng H, Chen X, Li T, Zhang J, Jin G, Cai D and Huang Z: FOXA1 is prognostic of triple negative breast cancers by transcriptionally suppressing SOD2 and IL6. Int J Biol Sci. 15:1030–1041. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Cao J, Wang X, Wang D, Ma R, Li X, Feng H, Wang J, Liu S and Wang L: PGC-1β cooperating with FOXA2 inhibits proliferation and migration of breast cancer cells. Cancer Cell Int. 19:932019. View Article : Google Scholar : PubMed/NCBI | |
|
Song Y, Zeng S, Zheng G, Chen D, Li P, Yang M, Luo K, Yin J, Gu Y, Zhang Z, et al: FOXO3a-driven miRNA signatures suppresses VEGF-A/NRP1 signaling and breast cancer metastasis. Oncogene. 40:777–790. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Wei G, Yang X, Lu H, Zhang L, Wei Y, Li H, Zhu M and Zhou X: Prognostic value and immunological role of FOXM1 in human solid tumors. Aging (Albany NY). 14:9128–9148. 2022. View Article : Google Scholar : PubMed/NCBI | |
|
Ha M and Kim VN: Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Broughton JP, Lovci MT, Huang JL, Yeo GW and Pasquinelli AE: Pairing beyond the seed supports microRNA targeting specificity. Mol Cell. 64:320–333. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Arora T, Kausar MA, Aboelnaga SM, Anwar S, Hussain MA, Sadaf S, Kaur S, Eisa AA, Shingatgeri VMM, Najm MZ and Aloliqi AA: miRNAs and the Hippo pathway in cancer: Exploring the therapeutic potential (Review). Oncol Rep. 48:1352022. View Article : Google Scholar : PubMed/NCBI | |
|
Denli AM, Tops BB, Plasterk RH, Ketting RF and Hannon GJ: Processing of primary microRNAs by the microprocessor complex. Nature. 432:231–235. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Alarcón CR, Lee H, Goodarzi H, Halberg N and Tavazoie SF: N6-methyladenosine marks primary microRNAs for processing. Nature. 519:482–485. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y and Tsukihara T: A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 326:1275–1279. 2009. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang H, Kolb FA, Jaskiewicz L, Westhof E and Filipowicz W: Single processing center models for human Dicer and bacterial RNase III. Cell. 118:57–68. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Wang W and Luo YP: MicroRNAs in breast cancer: Oncogene and tumor suppressors with clinical potential. J Zhejiang Univ Sci B. 16:18–31. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Corcoran C, Friel AM, Duffy MJ, Crown J and O'Driscoll L: Intracellular and extracellular microRNAs in breast cancer. Clin Chem. 57:18–32. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Guo X, Connick MC, Vanderhoof J, Ishak MA and Hartley RS: MicroRNA-16 modulates HuR regulation of cyclin E1 in breast cancer cells. Int J Mol Sci. 16:7112–7132. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Jin T, Suk Kim H, Ki Choi S, Hye Hwang E, Woo J, Suk Ryu H, Kim K, Moon A and Kyung Moon W: microRNA-200c/141 upregulates SerpinB2 to promote breast cancer cell metastasis and reduce patient survival. Oncotarget. 8:32769–32782. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Chen S, Wang Y, Ni C, Meng G and Sheng X: HLF/miR-132/TTK axis regulates cell proliferation, metastasis and radiosensitivity of glioma cells. Biomed Pharmacother. 83:898–904. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Wang D, Ren J, Ren H, Fu JL and Yu D: MicroRNA-132 suppresses cell proliferation in human breast cancer by directly targeting FOXA1. Acta Pharmacol Sin. 39:124–131. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Gao T, Zou M, Shen T and Duan S: Dysfunction of miR-802 in tumors. J Clin Lab Anal. 35:e239892021. View Article : Google Scholar : PubMed/NCBI | |
|
Yuan F and Wang W: MicroRNA-802 suppresses breast cancer proliferation through downregulation of FoxM1. Mol Med Rep. 12:4647–4651. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Wang Q, Ye B, Wang P, Yao F, Zhang C and Yu G: Overview of microRNA-199a regulation in cancer. Cancer Manag Res. 11:10327–10335. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Cuiffo BG, Campagne A, Bell GW, Lembo A, Orso F, Lien EC, Bhasin MK, Raimo M, Hanson SE, Marusyk A, et al: MSC-regulated microRNAs converge on the transcription factor FOXP2 and promote breast cancer metastasis. Cell Stem Cell. 15:762–774. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu C, Li J, Wang X and Song L: Unregulated miR-96 induces cell proliferation in human breast cancer by downregulating transcriptional factor FOXO3a. PLoS One. 5:e157972010. View Article : Google Scholar : PubMed/NCBI | |
|
Yin Z, Wang W, Qu G, Wang L, Wang X and Pan Q: MiRNA-96-5p impacts the progression of breast cancer through targeting FOXO3. Thorac Cancer. 11:956–963. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Tan X, Li Z, Ren S, Rezaei K, Pan Q, Goldstein AT, Macri CJ, Cao D, Brem RF and Fu SW: Dynamically decreased miR-671-5p expression is associated with oncogenic transformation and radiochemoresistance in breast cancer. Breast Cancer Res. 21:892019. View Article : Google Scholar : PubMed/NCBI | |
|
Kumar U, Hu Y, Masrour N, Castellanos-Uribe M, Harrod A, May ST, Ali S, Speirs V, Coombes RC and Yagüe E: MicroRNA-495/TGF-β/FOXC1 axis regulates multidrug resistance in metaplastic breast cancer cells. Biochem Pharmacol. 192:1146922021. View Article : Google Scholar : PubMed/NCBI | |
|
Badve S, Turbin D, Thorat MA, Morimiya A, Nielsen TO, Perou CM, Dunn S, Huntsman DG and Nakshatri H: FOXA1 expression in breast cancer-correlation with luminal subtype A and survival. Clin Cancer Res. 13:4415–4421. 2007. View Article : Google Scholar : PubMed/NCBI | |
|
Fu X, Jeselsohn R, Pereira R, Hollingsworth EF, Creighton CJ, Li F, Shea M, Nardone A, De Angelis C, Heiser LM, et al: FOXA1 overexpression mediates endocrine resistance by altering the ER transcriptome and IL-8 expression in ER-positive breast cancer. Proc Natl Acad Sci USA. 113:E6600–E6609. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D and Carroll JS: FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 43:27–33. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Yamaguchi N, Nakayama Y and Yamaguchi N: Down-regulation of Forkhead box protein A1 (FOXA1) leads to cancer stem cell-like properties in tamoxifen-resistant breast cancer cells through induction of interleukin-6. J Biol Chem. 292:8136–8148. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Karunarathna U, Kongsema M, Zona S, Gong C, Cabrera E, Gomes AR, Man EP, Khongkow P, Tsang JW, Khoo US, et al: OTUB1 inhibits the ubiquitination and degradation of FOXM1 in breast cancer and epirubicin resistance. Oncogene. 35:1433–1444. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Nestal de Moraes G, Delbue D, Silva KL, Robaina MC, Khongkow P, Gomes AR, Zona S, Crocamo S, Mencalha AL, Magalhães LM, et al: FOXM1 targets XIAP and Survivin to modulate breast cancer survival and chemoresistance. Cell Signal. 27:2496–2505. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Nestal de Moraes G, Bella L, Zona S, Burton MJ and Lam EW: Insights into a critical role of the FOXO3a-FOXM1 axis in DNA damage response and genotoxic drug resistance. Curr Drug Targets. 17:164–177. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Di Fruscia P, Zacharioudakis E, Liu C, Moniot S, Laohasinnarong S, Khongkow M, Harrison IF, Koltsida K, Reynolds CR, Schmidtkunz K, et al: The discovery of a highly selective 5,6,7,8-tetrahydrobenzo[4,5]thieno[2,3-d]pyrimidin-4(3H)-one SIRT2 inhibitor that is neuroprotective in an in vitro Parkinson's disease model. ChemMedChem. 10:69–82. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Saba R, Alsayed A, Zacny JP and Dudek AZ: The role of forkhead box protein M1 in breast cancer progression and resistance to therapy. Int Breast Cancer. 2016:97681832016.PubMed/NCBI | |
|
Ji X, Tian X, Feng S, Zhang L, Wang J, Guo R, Zhu Y, Yu X, Zhang Y, Du H, et al: Intermittent F-actin perturbations by magnetic fields inhibit breast cancer metastasis. Research (Wash DC). 6:00802023.PubMed/NCBI | |
|
Halasi M, Hitchinson B, Shah BN, Váraljai R, Khan I, Benevolenskaya EV, Gaponenko V, Arbiser JL and Gartel AL: Honokiol is a FOXM1 antagonist. Cell Death Dis. 9:842018. View Article : Google Scholar : PubMed/NCBI | |
|
Rajamanickam S, Panneerdoss S, Gorthi A, Timilsina S, Onyeagucha B, Kovalskyy D, Ivanov D, Hanes MA, Vadlamudi RK, Chen Y, et al: Inhibition of FoxM1-mediated DNA repair by imipramine blue suppresses breast cancer growth and metastasis. Clin Cancer Res. 22:3524–3536. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Lopez JS and Banerji U: Combine and conquer: Challenges for targeted therapy combinations in early phase trials. Nat Rev Clin Oncol. 14:57–66. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Martel S, Bruzzone M, Ceppi M, Maurer C, Ponde NF, Ferreira AR, Viglietti G, Del Mastro L, Prady C, de Azambuja E and Lambertini M: Risk of adverse events with the addition of targeted agents to endocrine therapy in patients with hormone receptor-positive metastatic breast cancer: A systematic review and meta-analysis. Cancer Treat Rev. 62:123–132. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Pegram MD, Konecny GE, O'Callaghan C, Beryt M, Pietras R and Slamon DJ: Rational combinations of trastuzumab with chemotherapeutic drugs used in the treatment of breast cancer. J Natl Cancer Inst. 96:739–749. 2004. View Article : Google Scholar : PubMed/NCBI | |
|
Waks AG and Winer EP: Breast cancer treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Guillen VS, Ziegler Y, Gopinath C, Kumar S, Dey P, Plotner BN, Dawson NZ, Kim SH, Katzenellenbogen JA and Katzenellenbogen BS: Effective combination treatments for breast cancer inhibition by FOXM1 inhibitors with other targeted cancer drugs. Breast Cancer Res Treat. 198:607–621. 2023. View Article : Google Scholar : PubMed/NCBI | |
|
Lin Z, Huang W, He Q, Li D, Wang Z, Feng Y, Liu D, Zhang T, Wang Y, Xie M, et al: FOXC1 promotes HCC proliferation and metastasis by Upregulating DNMT3B to induce DNA Hypermethylation of CTH promoter. J Exp Clin Cancer Res. 40:502021. View Article : Google Scholar : PubMed/NCBI | |
|
Bader AG and Lammers P: The therapeutic potential of microRNAs. Innov Pharm Technol. 52–55. 2011. | |
|
Broderick JA and Zamore PD: MicroRNA therapeutics. Gene Ther. 18:1104–1110. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Tate CR, Rhodes LV, Segar HC, Driver JL, Pounder FN, Burow ME and Collins-Burow BM: Targeting triple-negative breast cancer cells with the histone deacetylase inhibitor panobinostat. Breast Cancer Res. 14:R792012. View Article : Google Scholar : PubMed/NCBI | |
|
Linares A, Dalenc F, Balaguer P, Boulle N and Cavailles V: Manipulating protein acetylation in breast cancer: A promising approach in combination with hormonal therapies? J Biomed Biotechnol. 2011:8569852011. View Article : Google Scholar : PubMed/NCBI | |
|
Khafaga AF, Shamma RN, Abdeen A, Barakat AM, Noreldin AE, Elzoghby AO and Sallam MA: Celecoxib repurposing in cancer therapy: Molecular mechanisms and nanomedicine-based delivery technologies. Nanomedicine (Lond). 16:1691–1712. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
National Library of Medicine (NIH), . Study to Compare Alisertib with Paclitaxel vs Paclitaxel Alone in Metastatic or Locally Recurrent Breast Cancer. Clinical trial: NCT02187991. NIH; Bethesda, MD: 2022, https://www.mycancergenome.org/content/clinical_trials/NCT02187991/December 1–2022 | |
|
National Library of Medicine (NIH), . First Time in Human Study of AZD8701 With or Without Durvalumab in Participants with Advanced Solid Tumours. Clinical trial: NCT04504669. NIH; Bethesda, MD: 2022, https://www.mycancergenome.org/content/clinical_trials/NCT04504669/December 1–2022 | |
|
National Library of Medicine (NIH), . Pre-op Pembro + Radiation Therapy in Breast Cancer (P-RAD). Clinical trial: NCT04443348. NIH; Bethesda, MD: 2022, https://www.mycancergenome.org/content/clinical_trials/NCT04443348/December 1–2022 | |
|
National Library of Medicine (NIH), . A Study of PDR001 in Combination with LCL161, Everolimus or Panobinostat. Clinical trial: NCT02890069. NIH; Bethesda, MD: 2022, https://www.mycancergenome.org/content/clinical_trials/NCT02890069/December 1–2022 | |
|
National Library of Medicine (NIH), . Phase I, Dose Study to Look at the Safety and Pharmacokinetics of AZD8835 in Patients with Advanced Solid Tumours. Clinical trial: NCT02260661. NIH; Bethesda, MD: 2022, https://www.mycancergenome.org/content/clinical_trials/NCT02260661/December 1–2022 |