- 1Department of Oral Pathology, Radiology, and Medicine, College of Dentistry, University of Iowa, Iowa City, IA, United States
- 2Department of Pathology, College of Medicine, University of Iowa Hospitals and Clinics, Iowa City, IA, United States
- 3Department of Pathology, Microbiology and Immunology, Vanderbilt University Medical Center, Nashville, TN, United States
- 4Department of Otolaryngology – Head and Neck Surgery, University of Iowa Hospitals and Clinics, Iowa City, IA, United States
- 5Holden Comprehensive Cancer Center, University of Iowa Hospitals and Clinics, Iowa City, IA, United States
- 6Iowa City Veterans Affairs Health Care System, Iowa City, IA, United States
Background: Epidermal growth factor receptor (EGFR) is well known as a general prognostic biomarker for head and neck tumors, however the specific prognostic value of EGFR in oral squamous cell carcinoma (OSCC) is controversial. Recently, the presence of tumor-infiltrating T cells has been associated with significant survival advantages in a variety of disease sites. The present study will determine if the inclusion of T cell specific markers (CD3, CD4 and CD8) would enhance the prognostic value of EGFR in OSCCs.
Methods: Tissue microarrays containing 146 OSCC cases were analyzed for EGFR, CD3, CD4 and CD8 expression using immunohistochemical staining. EGFR and T cell expression scores were correlated with clinicopathological parameters and survival outcomes.
Results: Results showed that EGFR expression had no impact on overall survival (OS), but EGFR-positive (EGFR+) OSCC patients demonstrated significantly worse progression free survival (PFS) compared to EGFR-negative (EGFR-) patients. Patients with CD3, CD4 and CD8-positive tumors had significantly better OS compared to CD3, CD4 and CD8-negative patients respectively, but no impact on PFS. Combined EGFR+/CD3+ expression was associated with cases with no nodal involvement and significantly more favorable OS compared to EGFR+/CD3- expression. CD3 expression had no impact on OS or PFS in EGFR- patients. Combinations of EGFR/CD8 and EGFR/CD4 expression showed no significant differences in OS or PFS among the expression groups.
Conclusion: Altogether these results suggest that the expression of CD3+ tumor-infiltrating T cells can enhance the prognostic value of EGFR expression and warrants further investigation as prognostic biomarkers for OSCC.
Introduction
Oral cancer is the most common malignancy of the head and neck region (1, 2). More than 90% of all oral cancers are oral squamous cell carcinomas (OSCCs) (2) and arise in the tongue, floor of the mouth, palate, and labial and buccal mucosa (3). Despite the anatomical accessibility of OSCCs and advances in cancer diagnosis and treatment, the 5-year overall survival rate has remained at less than 50% for the last three decades (4). The main treatment approach for OSCC is surgery with post-operative radiotherapy (RT) with or without adjuvant systemic therapy (chemotherapy and/or targeted therapy). However, OSCC patients have a high risk of tumor recurrence and the main disease-related mortality in OSCC patients is due to locoregional failure (5). At this time, there are no prognostic or predictive molecular biomarkers used clinically for OSCCs. Treatment decisions depend primarily on tumor site, TNM classification, and clinicopathological parameters (6) which do not consistently predict patient prognosis (7, 8). There is an urgent need for more biomarker studies that will aid in rapid risk assessment and guide treatment decisions for OSCC patients
Epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase in the ErbB family of receptors (9, 10). EGFR is involved in multiple complex pathways in cell regulation including embryogenesis, tissue regeneration and homeostasis (11–13). Dysregulation of EGFR expression/signaling in oral epithelial cells can lead to the development and progression of OSCCs (14–18). As a result, EGFR when measured by quantitative and semiquantitative immunohistochemistry (IHC) (19–22), is generally known as an independent prognostic factor for tumor recurrence in OSCC patients. However, the prognostic accuracy of EGFR expression is unreliable and somewhat controversial due to the lack of EGFR expression in many tumors in the head and neck region (23) and a number of conflicting reports demonstrating no prognostic value (24–31). Because of this, EGFR expression is not routinely tested for in the clinical setting for OSCC (28, 31–36) despite its established role in tumor aggressiveness.
Recently, growing evidence for the interaction of tumor and immune cells in tumor growth, recurrence and progression has emerged (37–41). OSCCs due to its unique anatomical location are characterized by an abundant infiltrate of immune cells (42). However, depending on the make-up and location of the immune cells in the tumor microenvironment (TEM), tumor-supporting outcomes are possible that contribute to immune escape (43, 44). Cells that contribute to immune escape mechanisms include tumor-associated macrophages (45), regulatory dendritic cells (DCs) (46), T regulatory cells (Tregs) (47) and myeloid-derived suppressor cells (MDSCs) (48, 49). On the other hand, in many cancers including OSCCs, an active anti-tumor immune response is often reflected by the abundance of tumor-infiltrating lymphocytes (TILs) such as CD8+ T cells (41, 50, 51) and is correlated with favorable prognosis (37–40). Tumor-infiltrating CD4+ T cells have recently been correlated with favorable prognosis in OSCCs (52) however CD4+ T cells consist of several subpopulations (including Tregs) and CD4 expression as a prognostic biomarker in OSCCs and HNSCCs is controversial (40, 53–55). Together the assessment of TILs is promising as a prognostic tool for OSCCs.
Given that the presence of TILs provides important information regarding overall prognosis in OSCCS, and EGFR expression provides valuable information regarding the risk of tumor recurrence/progression, the goal of this study is to investigate if TIL expression would enhance the prognostic value of EGFR in OSCC patients.
Materials and Methods
Tissue Microarrays
Formalin-fixed paraffin-embedded tumor samples of patients were obtained from 266 surgical resection specimens of OSCC spanning 10 years of time (2005-2014) from the archives of the Department of Pathology at the University of Iowa Hospitals and Clinics. Cases were chosen selectively to ensure patients with no history of radiation or chemotherapy and to ensure a mixture of patients regarding recurrent status, node metastases, margin status, age, and smoking status. After excluding cases with unavailable tissue blocks, 146 cases were included in the current study. TMAs were constructed using 3-6 morphologically representative areas (tumor and stroma). Sections (4μm) were obtained from the TMAs on poly-L-lysine-coated glass slides. Routine hematoxylin-and eosin (H&E) sections were reviewed to confirm the original diagnosis. Subject clinicopathological characteristics considered were age, sex, smoking history, tumor site, T stage, N stage, differentiation, and presence of perineural invasion, lymphovascular invasion, bone invasion and local recurrence. Clinicopathological characteristics were obtained from medical records where tumor microscopic features and TNM stage had been previously evaluated by board certified pathologists. The TNM status was based on the American Joint Committee on Cancer 7 (AJCC7).
Immunohistochemistry
Antigen retrieval was performed on freshly cut sections in a decloaking chamber for 5 min at 125°C in TRIS buffer (pH 9.0). Endogenous peroxidase was blocked by incubation with 3% peroxide at room temperature for 8 min. IHC was performed with the following antibodies: EGFR (H11, Dako) at 1:200 dilution, CD3 (Dako A0452) at 1:200 dilution, CD4 (Novocastra NCL-L-CD4-368) at 1:100 dilution, and CD8 (Dako M7103) at 1:100 dilution. Bound antibody was detected using the HRP-DAB Cell & Tissue Staining Kit. All slides were counterstained with hematoxylin.
Quantification of EGFR and TIL Staining:
EGFR immunostaining was evaluated by semiquantitative scoring (score 0-3) where 0 represents staining in <10% neoplastic cells and 1, 2, and 3 representing weak, moderate, and strong staining in >10% neoplastic cells according to Gamboa-Domingez and colleagues (56). Immunostaining scores of 3 and 2 were designated as EGFR-positive (EGFR+) and scores of 1 and 0 were designated as EGFR-negative (EGFR-). CD3+, CD8+, CD4+ and FoxP3+ TILs were evaluated based on the density of positive inflammatory cells (score 0-3): 0 represents none or very few positive cells; 1 represents single isolated positive cell or small aggregates of positive cells 2-4 cells; 2 represents discrete nodules or aggregates of positive cells more than 2-4 cells; 3 represents bands or continuous aggregates of positive cells. For CD3, the immunostaining scores of 3 and 2 were designated as high CD3 (CD3+), while scores of 1 and 0 were designated as low CD3+ (CD3-). Due to low number of specimens that received scores of 3 (n=2) and 2 (n=11) for CD8, the immunostaining scores of 3, 2 and 1 were designated as CD8+, while a score of 0 were designated as CD8-. For CD4, immunostaining scores of 3 and 2 were designated as high CD4 (CD4+), while scores of 1 and 0 were designated as low CD4 (CD4-). Immunoexpression of EGFR, CD3, CD4 and CD8 was scored by 2 pathologists. In the event of disagreement, a consensus score was given. Each tissue core received an individual score, and the average score was calculated from all the tissue cores that were from the same case and used as a final score. Very few HPV+ OSCC cases were present in the patient cohort and were excluded from the study.
Statistical Analysis
Power for a sample size of 146 cases was estimated at 95.2% calculating from previous work (57). The association between expression scores and patient clinicopathological characteristics were analyzed by Chi-square test. Survival outcomes differences were plotted using the Kaplan-Meier method while estimates for the group hazard ratios were obtained using Cox proportional hazards (PH) modeling. Overall survival (OS) is defined as the length of time (in months) from the date of diagnosis that the patients remain alive. Progression free survival (PFS) is defined as the time from diagnosis to disease progression or death (in months) from any cause. All testing was performed on the univariate level and unadjusted for multiple comparisons. Differences between survival curves were compared using the log-rank test. A p-value below 0.05 was considered statistically significant. GraphPad Prism 8.1.2 was utilized for data analysis.
Results
Clinicopathological Characteristics
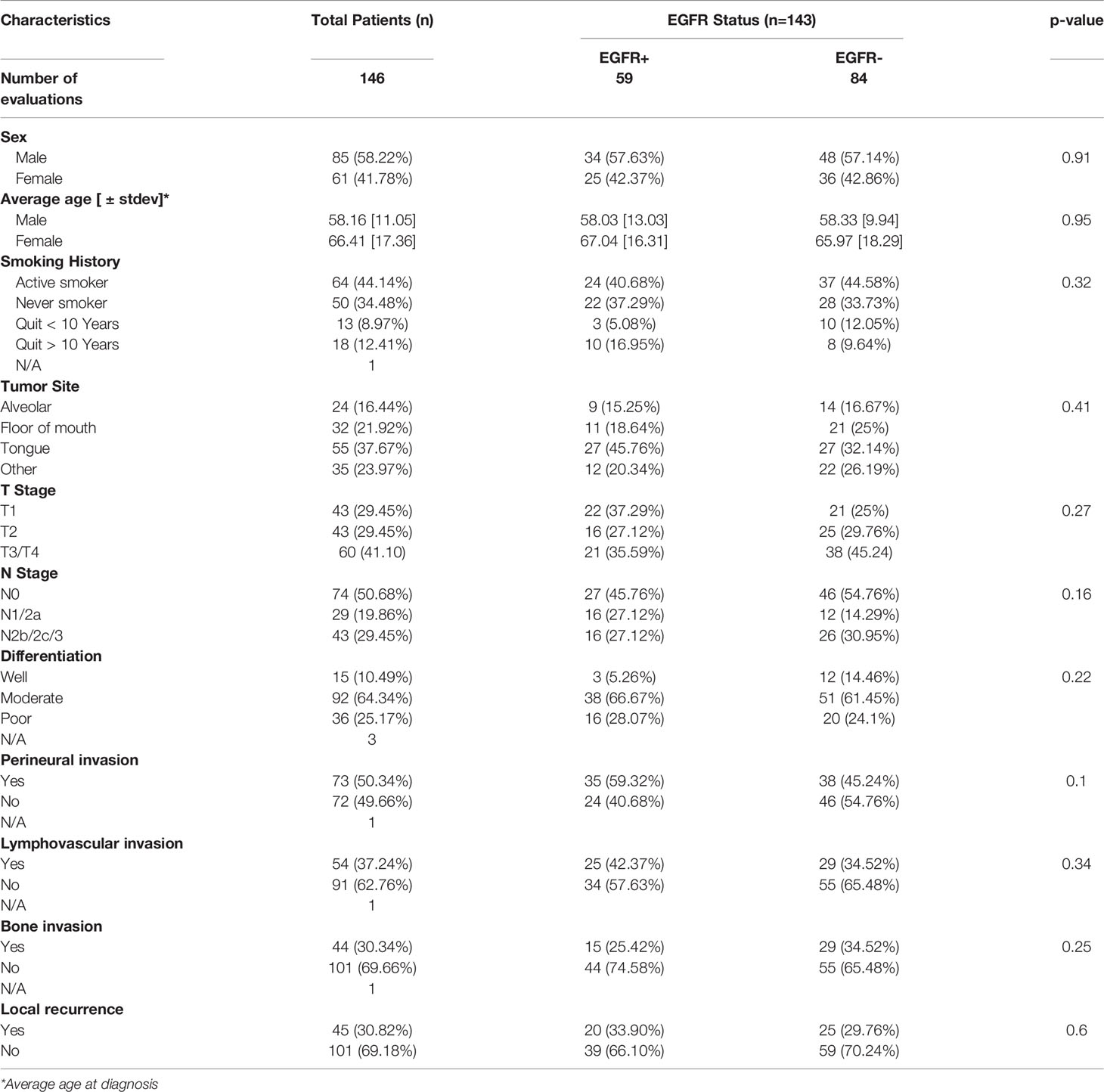
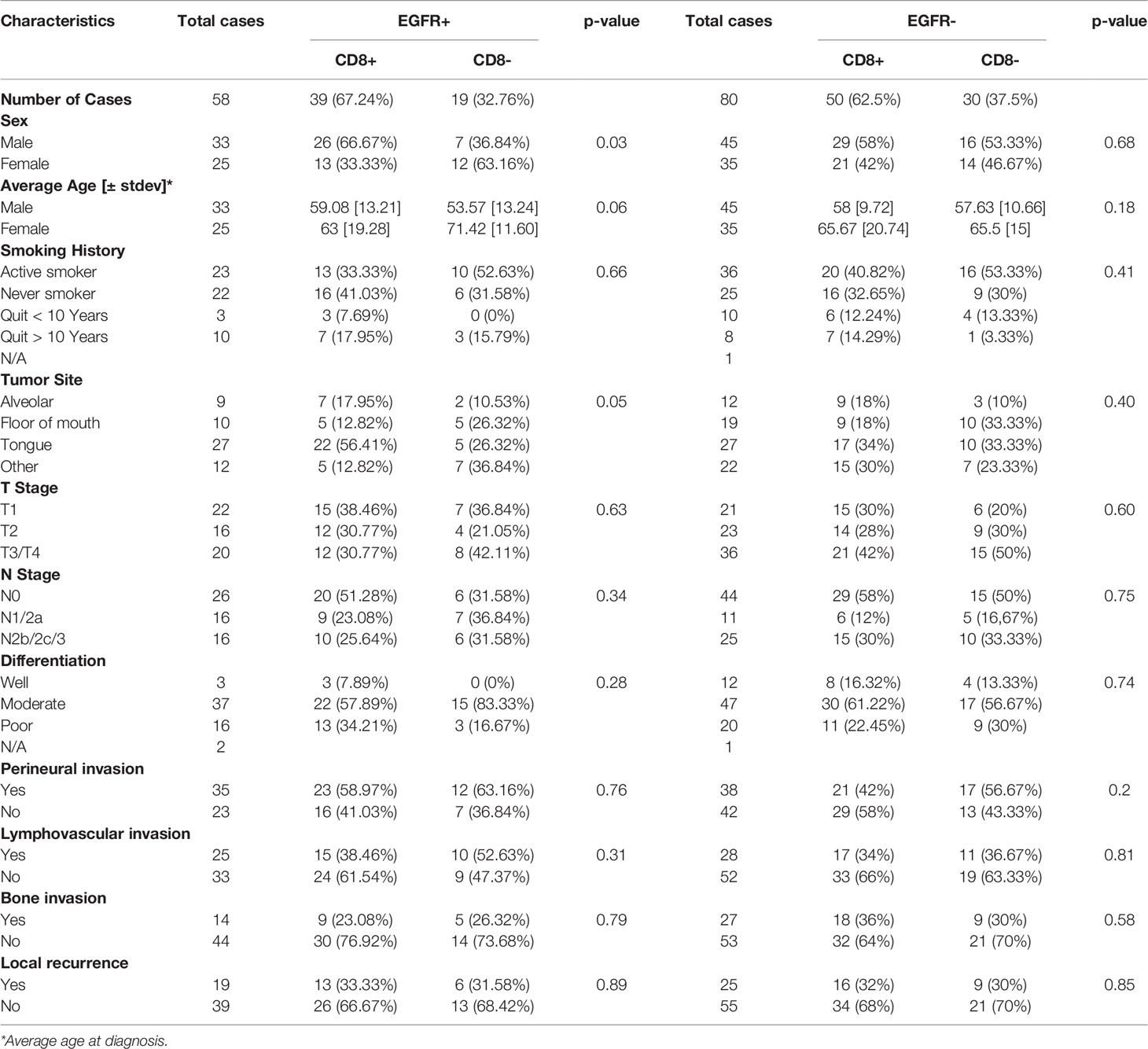
Baseline characteristics for the OSCC patients are summarized in Table 1. Fifty eight percent of the patients were male and 42% were female. Female patients were diagnosed at an older average age then male patients (66 versus 58 years respectively, p=0.002, Table 1), although there were no significant differences in survival outcomes (OS or PFS) between sexes (Supplemental Figures 1A, B). Patients 60 years and older demonstrated significantly worse OS (but not PFS) compared to patients under 60 years (Figures 1A, B). Active smokers comprised 44% of patients, with 35% having never smoked, 9% which quit smoking for less than 10 years, and 12% which quit smoking for more than 10 years (Table 1). There was no difference in OS between active smokers, never smokers and patients that quit (p=0.2, Figure 1C), however significant differences were observed in PFS where never smokers had improved PFS compares to active and patients that quit smoking (p=0.04, Figure 1D). Of the oral cavity disease sites, tongue was the most common disease location (38%, Table 1) and there was a trend toward differences in OS (p=0.07) but not PFS (p=0.38) by tumor site (Supplemental Figures 1C, D). Patients with T3/T4 tumors represented 41% of the patient cohort (Table 1) and showed a trend toward worse OS compared to T1 and T2 tumors (p=0.06) but no differences in PFS (Supplemental Figures 1E, F). The majority of patients presented with no lymph node metastasis (N0, 51%, Table 1) and demonstrated significantly more favorable OS (but not PFS) compared to patients that presented with N1 or N2/N3 disease (Figures 2A, B). Patients with poorly differentiated tumors represented 25% of the patient cohort (Table 1) and were associated with a trend toward worse OS (p=0.08, Figure 2C) and significantly worse PFS (p=0.04, Figure 2D) compared to well and moderately differentiated tumors. Perineural (PNI), lymphovascular (LVI) and bone invasion (BI) were observed in 50%, 37% and 30% of tumors respectively (Table 1). Patients presenting with PNI demonstrated significantly worse OS (p=0.004, Figure 2E) and PFS (p=0.04, Figure 2F) compared to patients with no PNI. Patients presenting with LVI demonstrated significantly worse OS (p=0.004, Figure 2G) and a trend toward worse PFS (p=0.06, Figure 2H) compared to patients with no LVI. BI status did not affect OS nor PFS in the patient cohort (Supplemental Figures 1G, H). The average follow-up time was 107 ± 37 months. Of the 146 cases, 48 patients survived 5 years after diagnoses.
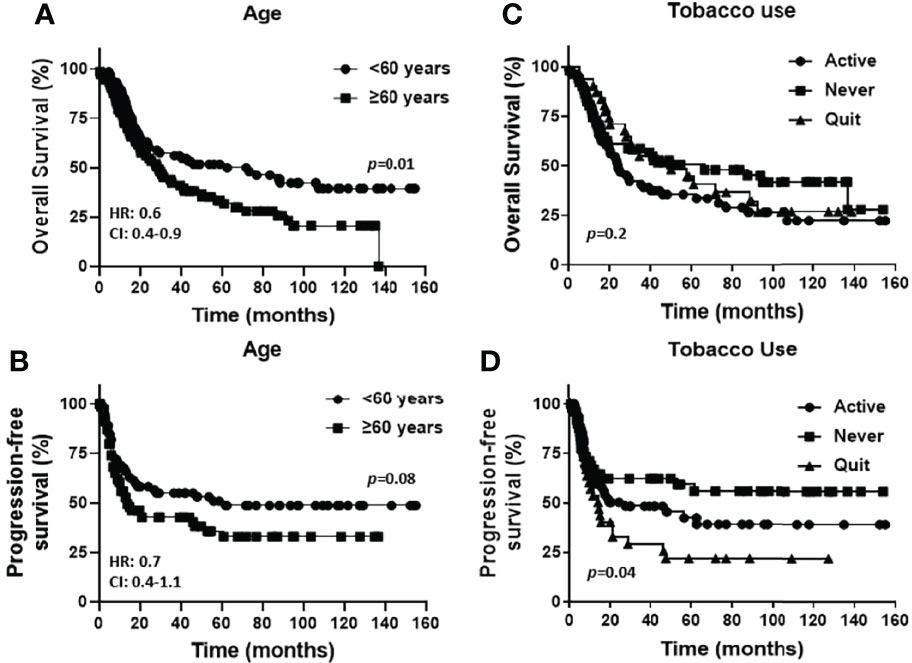
Figure 1 Prognostic impact of age and smoking history in OSCCs. Shown are Kaplan-Meier estimates of the overall survival (A, C) and progression free survival (B, D) of OSCC patients stratified by age [< 60 years or ≥ 60 years, (A, B)] and tobacco use history [active, never or quit, (C, D)]. HR: hazard ratio, CI: 95% confidence interval.
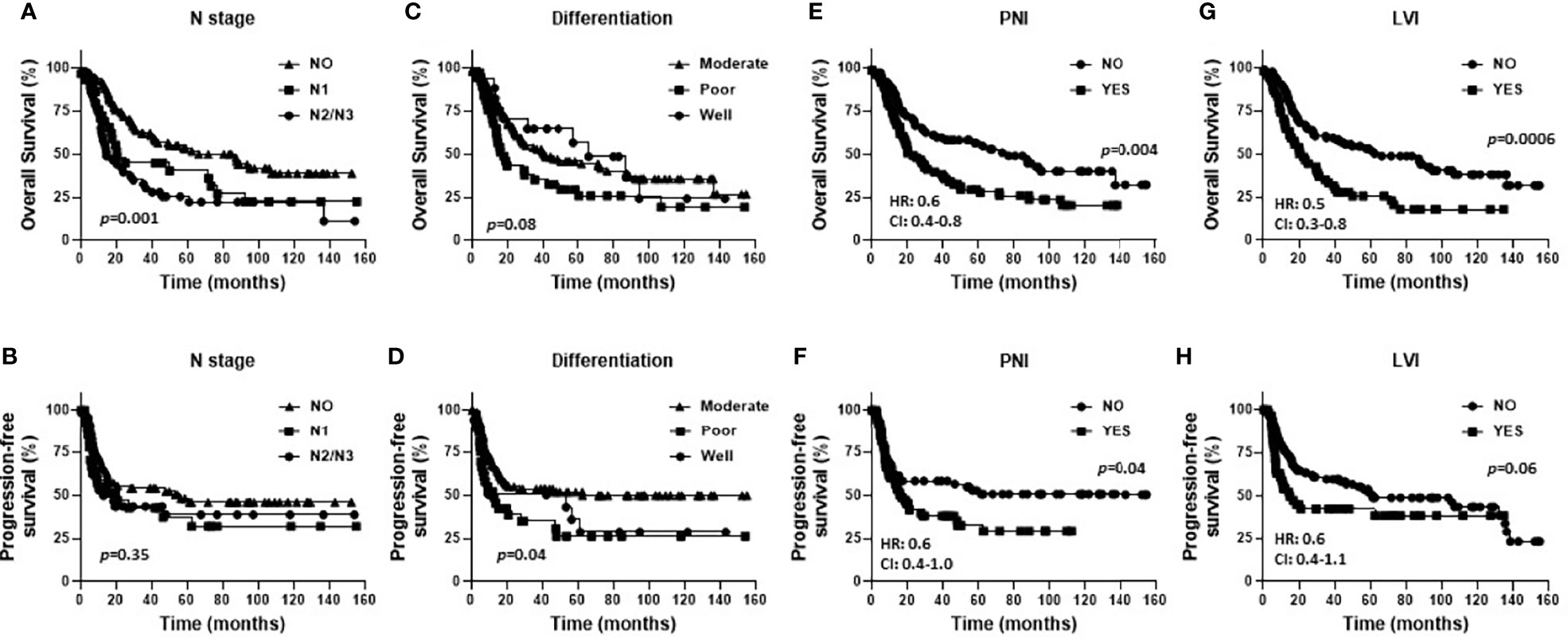
Figure 2 Prognostic impact of pathological factors in OSCCs. Shown are Kaplan-Meier estimates of overall survival (A, C, E, G) and progression free survival (B, D, F, H) of OSCC patients stratified by N stage (A, B), differentiation (C, D), presence of perineural invasion (PNI) (E, F), and presence of lymphovascular invasion (LVI) (G, H). HR, hazard ratio; CI, 95% confidence interval.
Prognostic Impact by EGFR Expression
The prognostic value of EGFR expression in the OSCC cases represented in the TMA was initially evaluated. Examples of EGFR expression scores are shown in Figure 3A. EGFR+ expression represents tumors with strong (score of 3) and moderate (score of 2) expression, while EGFR- expression represents low (score of 1) and no (score of 0) expression. Patient clinicopathological characteristics based on EGFR status (EGFR+ vs EGFR-) are shown in Table 1 where there were no significant associations observed. There were also no differences observed in OS (p=0.14) according to EGFR+ and EGFR- expression (Figure 3B). However significant differences were observed in PFS (p=0.01) with higher EGFR expression (EGFR+) being associated with worse PFS (Figure 3C). Similar results with OS and PFS were obtained when the prognostic value of each EGFR expression score (3, 2, 1 and 0) was previously evaluated shown here (58). However, the combined (3 + 2 [EGFR+], 1 + 0 [EGFR-]) expression scores were utilized for the remainder of the study to maintain sufficient case numbers to further analyze T cell subsets in these EGFR expression groups. These results support prior reports that EGFR expression is a strong predictor for PFS but not OS in OSCC patients (27, 59).
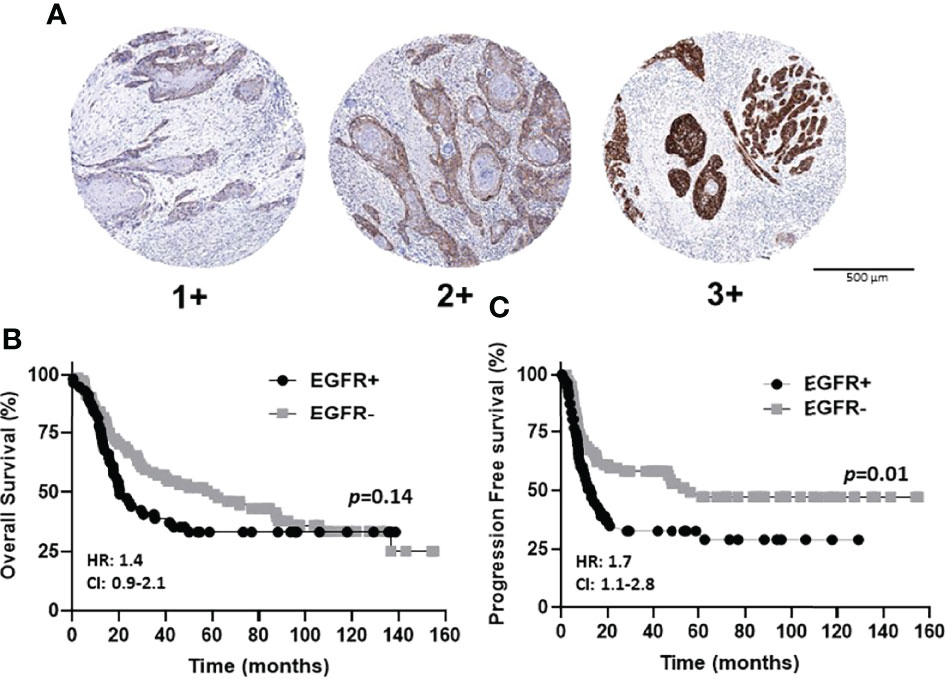
Figure 3 EGFR immunostaining and expression scores in OSCCs. (A): Shown are images of low [1+], moderate [2+], and strong [3+] EGFR expression scores. (B, C): Shown are Kaplan-Meier estimates of the overall survival (B) and progression free survival (C) of EGFR+ (2+ and 3+) and EGFR- (0 and 1+) patients. HR, hazard ratio; CI, 95% confidence interval.
Prognostic Impact of CD3+ Tumor-Infiltrating Lymphocyte Marker Expression
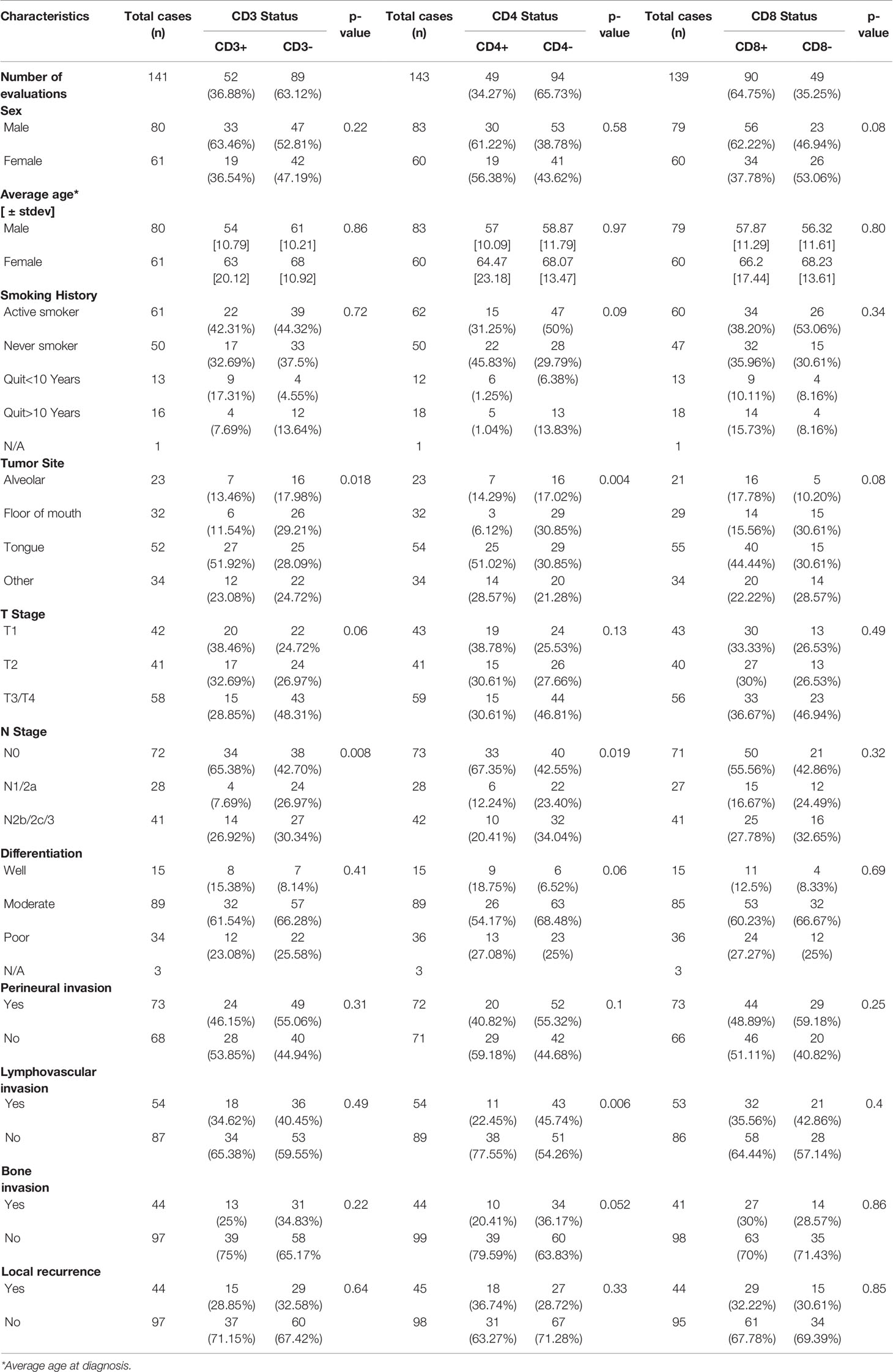
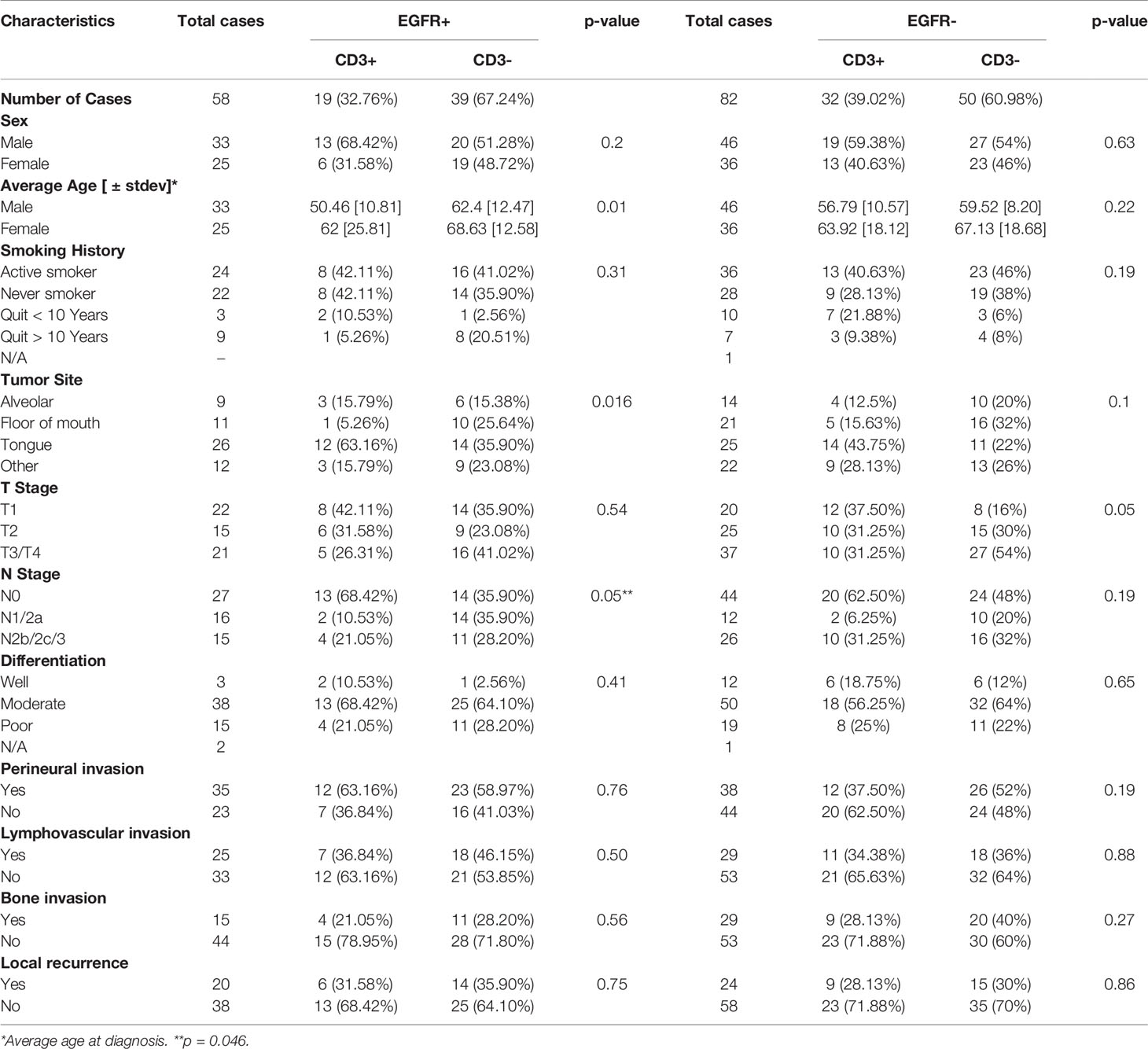
The prognostic value of TIL expression was initially assessed by pan-T cell (CD3) expression. Images of no [0], low [1+], moderate [2+], and strong [3+] CD3 expression scores are shown in Figure 4A. CD3 expression scores were combined in a manner identical to EGFR above where combined 3 and 2 scores represent CD3+ and combined 1 and 0 scores represented CD3-. CD3+ expression was associated with tongue tumors compared to other disease sites (p=0.018), and also associated with cases presenting with no lymph node metastasis (N0) compared to N1 and N2/3 cases (p=0.008) (Table 2). Additionally, CD3+ OSCCs were associated with favorable OS compared to CD3- tumors (p=0.01, Figure 4B). CD3 expression did not impact PFS (p=0.19, Figure 4C). These results suggest that CD3+ TILs may suppress lymph node metastasis and thus survival outcomes. The prognostic value of CD3 was next evaluated in EGFR+ and EGFR- OSCC patients. There was no difference in CD3 expression between EGFR+ and EGFR- tumors (Figure 5A). However, EGFR+/CD3+ expression was most frequently observed in younger males (p=0.01) and associated with less lymph node metastasis (p=0.046) compared to EGFR+/CD3- patients (Table 3). Patients with CD3+ OSCCs were associated with significantly higher OS (p=0.02, cox ph = 0.0097) compared to CD3- tumors, but only in EGFR+ patients (Figure 5B) and not EGFR- patients (Figure 5C). There were no differences in PFS observed with any of the combined EGFR/CD3 expression scores (Supplemental Figures 2A, B).
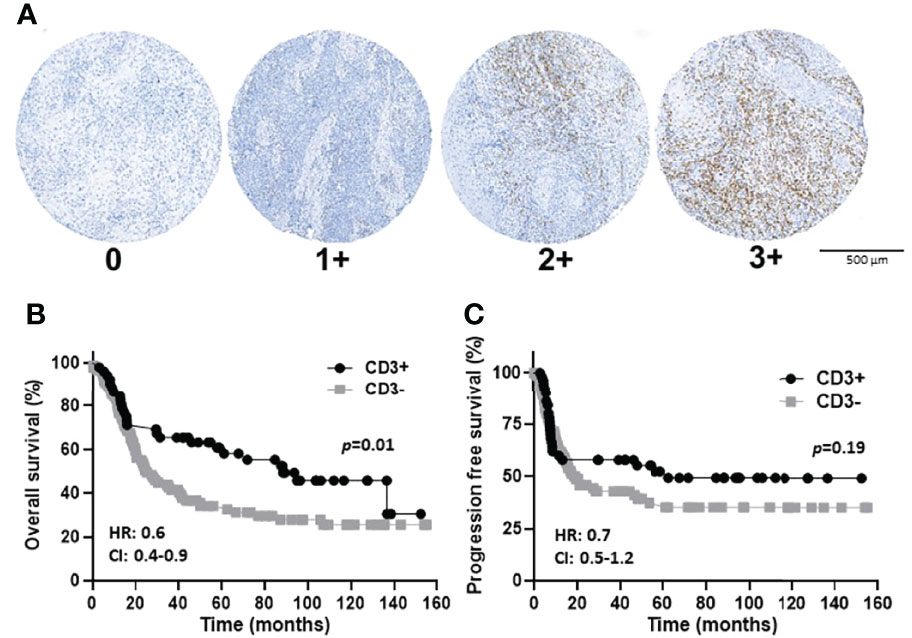
Figure 4 CD3 immunostaining and expression scores in OSCCs. (A): Shown are images of no [0], low [1+], moderate [2+], and strong [3+] CD3 expression scores. (B, C): Shown are Kaplan-Meier estimates of the overall survival (B) and progression free survival (C) of CD3+ (2+ and 3+) and CD3- (0 and 1+) patients. HR, hazard ratio; CI: 95% confidence interval.
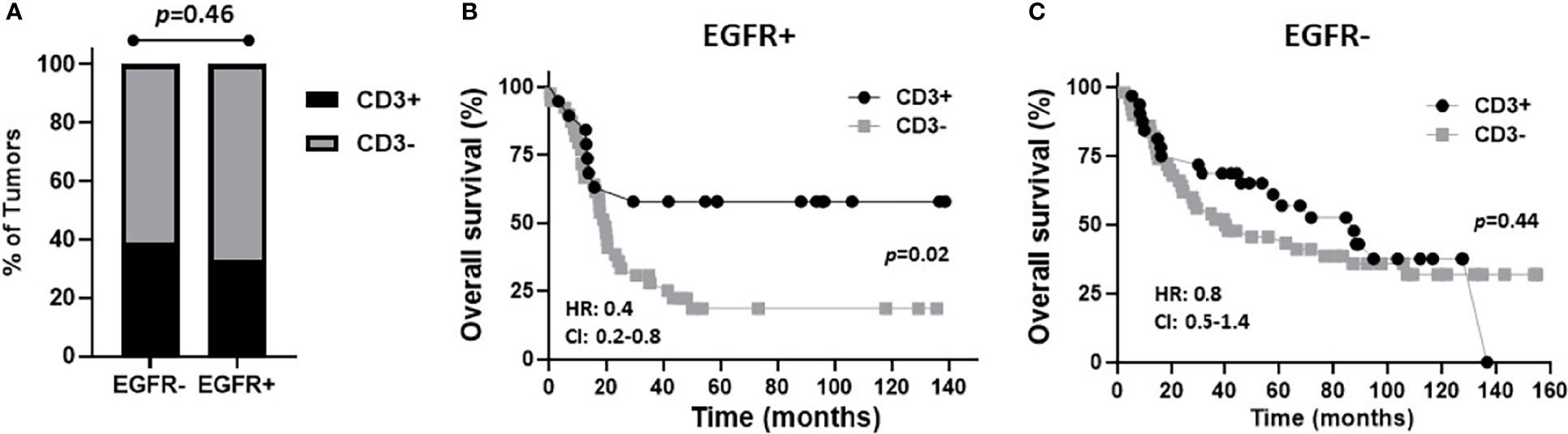
Figure 5 Prognostic impact of combined EGFR and CD3 expression in OSCCs. (A): Shown are percentages of tumors with CD3+ and CD3- expression based on EGFR status. (B, C): Shown are Kaplan-Meier estimates of overall survival of CD3+ and CD3- patients based on EGFR+ (B) or EGFR- (C) tumor expression. HR, hazard ratio; CI: 95% confidence interval.
Prognostic Impact of CD4+ and CD8+ Tumor-Infiltrating Lymphocyte Marker Expression
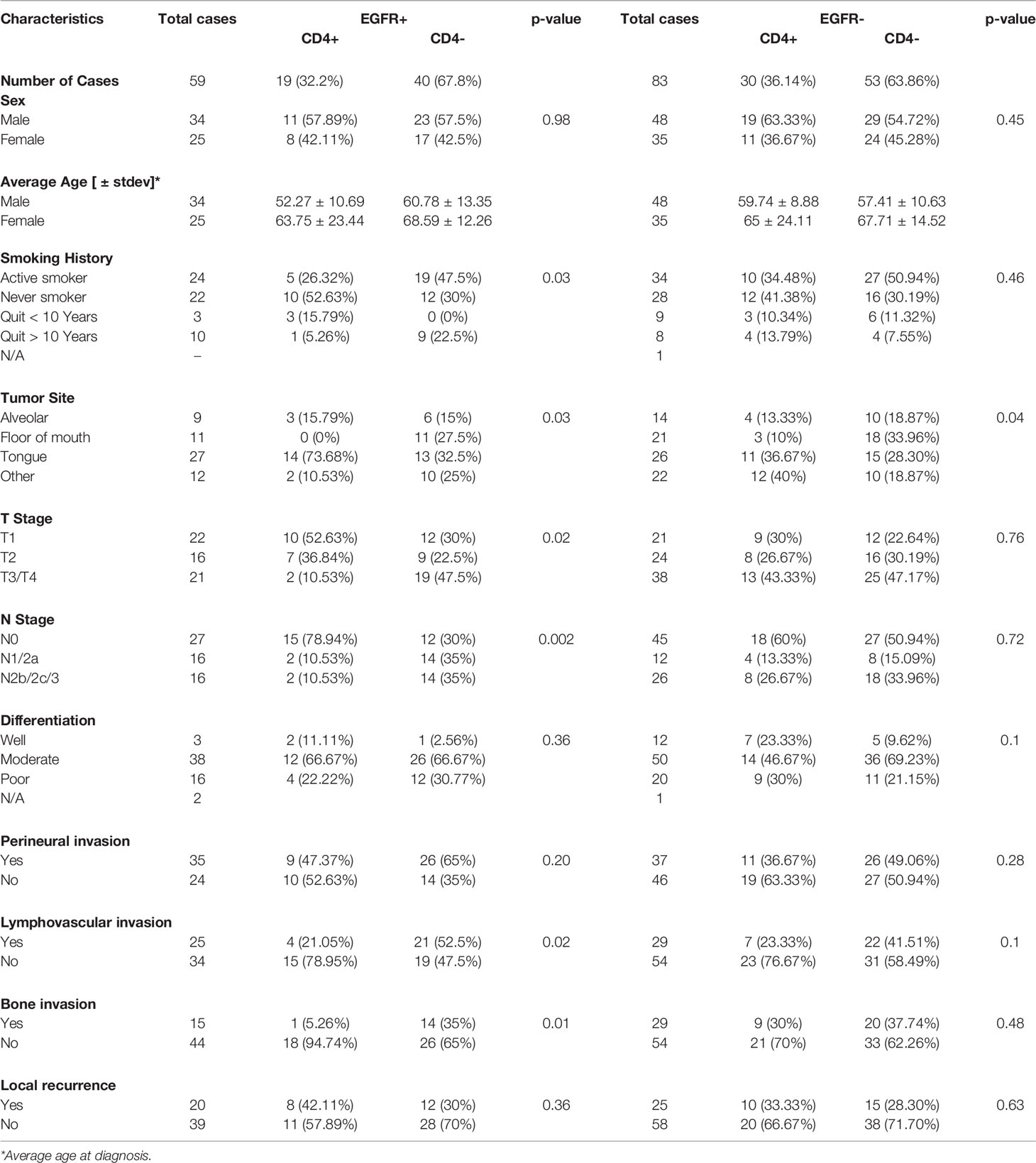
In order to determine what type of TIL represented the CD3 expression observed in the EGFR+/CD3+ patients, CD4 and CD8 expression was assessed in each OSCC case. CD4 expression scores and images are shown in Figure 6A and were combined as carried out for EGFR and CD3 where 3 and 2 scores represented CD4+ and combined 1 and 0 scores represented CD4-. Patient clinicopathological characteristics based on CD4 expression are shown in Table 2. CD4+ expression was associated with tongue tumors, no lymph node metastasis (N0, p=0.019) and no LVI (p=0.006, Table 2). CD4- expression were associated with floor of mouth tumors (p=0.004, Table 2). CD4- OSCCs were associated with significantly lower OS compared to CD4+ OSCCs (p=0.048, Figure 6B) and there was no impact on PFS (Supplemental Figure 3A). When combined EGFR and CD4 expression was analyzed, it was found that EGFR+/CD4+ expression was associated with never smokers (p=0.03), tongue tumor disease site (p=0.03), T1 tumors (p=0.02), N0 tumors (p=0.002), no LVI (p=0.02) and no BI (p=0.01) (Table 4). There was no difference in CD4 expression between EGFR+ and EGFR- tumors (p=0.69, Figure 6C) and there were no differences observed in OS (Figures 6D, E) or PFS (Supplemental Figures 3B, C) with any of the combined EGFR/CD4 expression scores. Patients that were CD3+ also were more likely to be CD4+, but EGFR status had no influence on this observation (Figure 6F). These results suggest that combined EGFR+/CD4+ expression is associated with favorable clinicopathological features for survival.
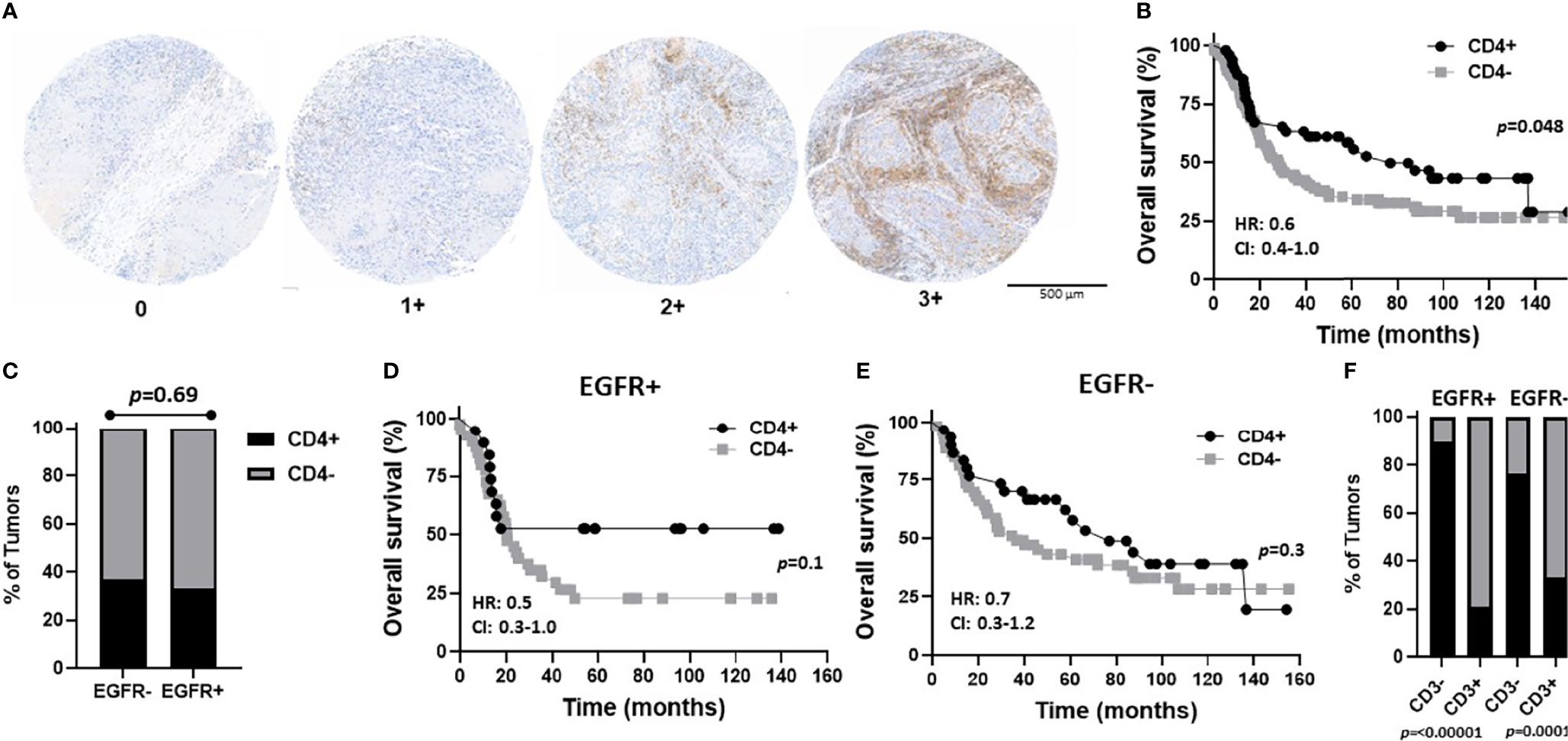
Figure 6 Prognostic impact of combined EGFR and CD4 expression in OSCCs. (A) Shown are images of no [0], low [1+], moderate [2+], and strong [3+] CD4 expression scores. (B) Kaplan-Meier estimates of the overall survival of CD4+ (2+, 3+) and CD4- (0, 1+) patients. (C) Percentages of tumors with CD4+ and CD4- expression based on EGFR status. (D, E) Kaplan-Meier estimates of overall survival of CD4+ and CD4- patients based on EGFR+ (D) or EGFR- (E) tumor expression. (F) Percentages of tumors with CD4+ and CD4- expression based on EGFR and CD3 status. HR, hazard ratio; CI, 95% confidence interval.
To assess CD8 expression, scores of 0-3 were again assigned as shown in Figure 7A. However, due to the low number of patients with CD8 scores of 3 (n=2) and 2 (n=11), we used scores of 3, 2, and 1 to represent CD8+ tumors (n=90) and a score of 0 to represent CD8- tumors (n=49) in order to have sufficient numbers for group comparisons and further subset analyses. Patient clinicopathological characteristics based on CD8 expression are included in Table 2 and there was no significant association between CD8 expression and any of the characteristics analyzed. However, CD8+ OSCCs were associated with significantly more favorable OS compared to CD8- OSCCs (p=0.02, Figure 7B) and there was no impact on PFS (Supplemental Figure 3D). When combined EGFR and CD8 expression was analyzed, EGFR+/CD8+ was associated with tumors in male patients but no other associations with clinicopathological characteristics were observed (Table 5). There were no differences in CD8 expression between EGFR+ and EGFR- tumors (p=0.66, Figure 7C). There were also no differences observed in OS (Figures 7D, E) or PFS (Supplemental Figures 3E, F) with any of the combined EGFR/CD8 expression scores. Notably, EGFR-/CD8+ expression showed a trend (p=0.07) toward association with favorable OS compared with EGFR-/CD8- expression when analyzed by log-rank test (Figure 7E), but showed significance (p=0.01) when the Gehan-Breslow-Wilcoxon test was used which gives more weight to deaths that occur at early time points. As shown with CD4+ (Figure 6F), patients that were CD3+ were also more likely to be CD8+ but EGFR status had no influence on this observation (Figure 7F).
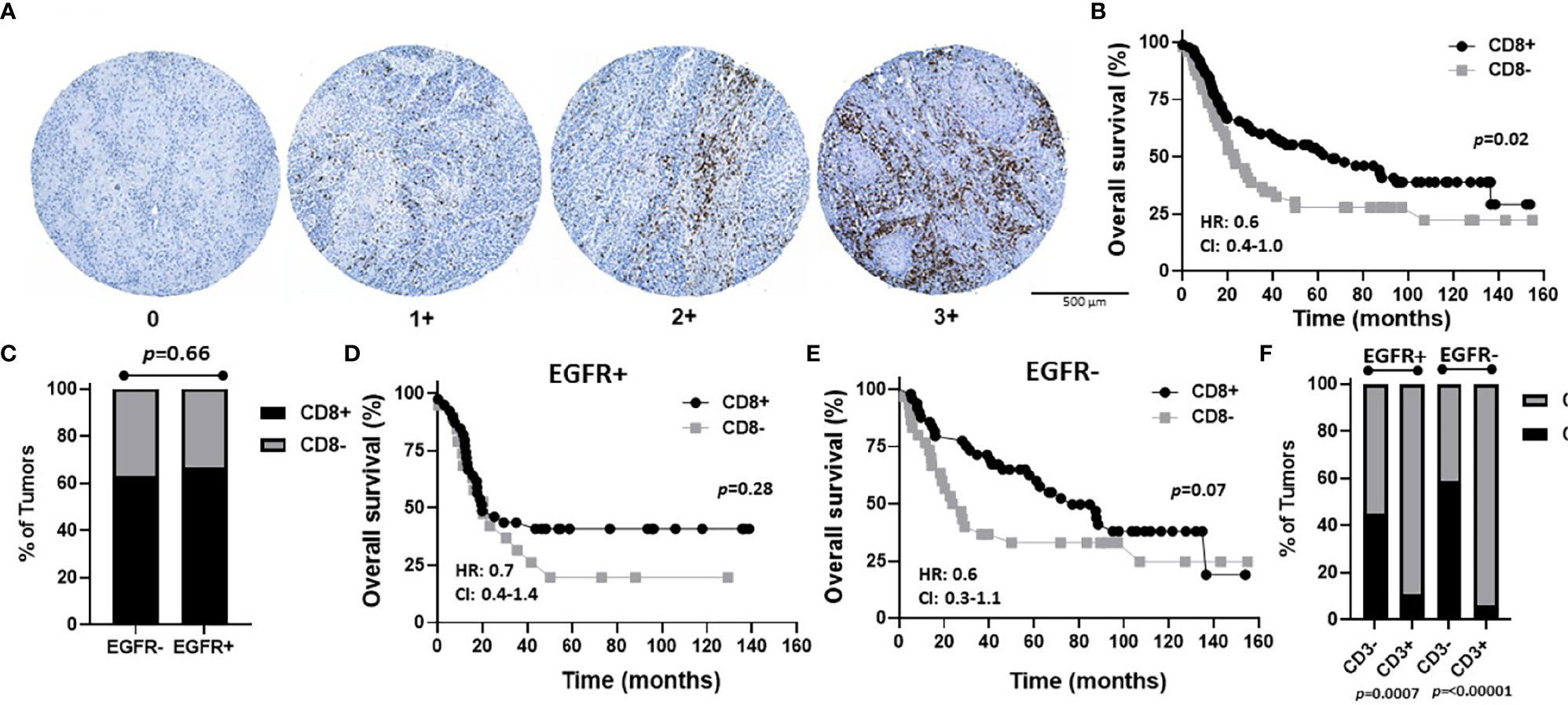
Figure 7 Prognostic impact of combined EGFR and CD8 expression in OSCCs. (A) Shown are images of no [0], low [1+], moderate [2+], and strong [3+] CD8 expression scores. (B) Kaplan-Meier estimates of the overall survival of CD8+ (2+, 3+, 1+) and CD8- (0) patients. (C) Percentages of tumors with CD8+ and CD8- expression based on EGFR status. (D, E) Kaplan-Meier estimates of overall survival of CD8+ and CD8- patients based on EGFR+ (D) or EGFR- (E) tumor expression. (F) Percentages of tumors with CD8+ and CD8- expression based on EGFR and CD3 status. HR, hazard ratio; CI, 95% confidence interval.
Altogether, these results demonstrate that combined EGFR+/CD3+ expression identified a subset of OSCC patients with favorable prognosis compared to other biomarker profiles which may be attributed to a lack of lymph node metastasis.
Discussion
The data shown here suggests that CD3 positivity is a prognostic biomarker for OSCC patients (Figure 4B) but only for EGFR+ tumors (Figure 5B). Hence, a combined EGFR/CD3 expression profile could potentially be used to make important treatment related decisions for EGFR+ patients that typically have an increased likelihood of tumor recurrence and progression (Figure 3C). For example, agents that re-direct T cells toward EGFR+ tumors such as EGFR-CD3 bispecific antibodies (60), can be administered to EGFR+/CD3- patients. Alternatively, agents that enhance T cell activity such as checkpoint inhibitors, can be administered to EGFR+/CD3+ patients.
Unfortunately, EGFR expression is not routinely tested for in the clinical setting for OSCC due to (1) EGFR expression not necessarily correlating with EGFR activity, (2) EGFR expression not predicting response to EGFR inhibitors, and (3) cetuximab (CTX, EGFR inhibitor) being administered to patients despite EGFR expression (24–26). This would also explain why CTX-based therapy has only modest impact on OSCC patients and in most cases is being replaced with immunotherapy (61). However, these therapy-related issues related to EGFR do not negate the important role of EGFR signaling in tumor growth and aggressiveness or the clear role of EGFR as a prognostic biomarker for PFS (24, 7).
As immunotherapy has now moved to the forefront of cancer therapy, immune biomarkers are of great interest. The expression of T cell markers can give an idea of the status of the immune microenvironment and possibly host immune responses (41). Increased expression and/or density of CD3 (pan-T cells) and CD8 (cytotoxic T cells) have previously shown associations with OS in a variety of disease models (37–40) which we also confirmed in the present studies (Figures 4B, 7B). The prognostic role of CD4 in the research literature is questionable (39, 41, 62, 63), although our studies found a significant prognostic role of CD4 in OS (Figure 6B). Among the OSCC locations, floor of the mouth tumors were more likely to CD4- and tumors of the tongue were more likely to express higher levels of T cell markers (especially CD3 and CD4) compared to the other oral cavity subsites. CD4+ expression was also associated with an absence of LVI (Table 2). We are unclear of why these TIL differences in oral cavity subsites occur, but we can reason that due to its location and function in the oral cavity, the tongue is constantly challenged by antigens from food and air. It is possible that the tongue (and tumors derived from the tongue) are T cell rich due to the abundance of T-cells residing in the mucosa that normally control mucosal immunity and tolerance (64). Nevertheless, our findings support prior work which found that most of tongue tumors are “TIL-high” (65, 66), the TILs were associated with absence of LVI (67) and were likely CD4+ T cells (68).
In this study we probed if EGFR could be combined with a TIL marker with the rationale that combining independent prognostic markers may increase the accuracy or reliability of assessing survival outcomes. EGFR expression would provide information about tumor aggressiveness while TIL marker(s) expression would give some input regarding the tumor immune microenvironment and overall prognosis. We found that combined EGFR/CD3 expression provided important prognostic information where increased CD3 expression was associated with improved OS in EGFR+ patients (Figure 5B). These results were surprising since we initially proposed based off individual EGFR (Figure 3B) and CD3 (Figure 4B) survival curves, that an EGFR-/CD3+ expression profile would be associated with the most favorable survival outcomes. However, CD3 expression provided no prognostic value in EGFR- tumors (Figure 5C). Perhaps since EGFR is a self-antigen, increased EGFR expression may trigger an EGFR-specific T cell response which would explain why EGFR+/CD3+ patients have a favorable OS. In support of this idea, previous work has shown that EGFR expressed on HNSCC cells induces a specific immune response in vivo and that higher EGFR expression was associated with increased circulation of EGFR-specific CD8+ T cells (69).
The majority of patients with high CD3 expression were high for CD4 (Figure 6F) and CD8 (Figure 7F) expression regardless of EGFR expression. Therefore, it is difficult to assess if increased CD4+ and/or CD8+ T cell activity is responsible for the favorable OS observed in EGFR+/CD3+ patients. Interestingly, combined EGFR+/CD4+ expression was associated with a variety of characteristics associated for favorable survival such as history of not smoking, T1 and N0 stage, and absence of LVI and BI (Table 4). However, EGFR+/CD4+ expression was not associated with OS or PFS despite these findings (Figures 6D,E). Future work will pursue these observations and further determine the expression of subsets of CD4+ T cells in OSCCs combined with EGFR. We expected to observe superior survival outcomes in EGFR-/CD8+ patients since EGFR- and CD8+ expression as separate entities are associated with more favorable outcomes compared to EGFR+ and CD8- respectively. In support of this, we found a strong trend (p=0.07) between combined EGFR-/CD8+ expression and survival outcomes (Figure 7E). The sole explanation for the favorable OS observed in EGFR+/CD3+ patients was the significant association with decreased lymph node metastasis (Table 2). This finding is quite contradictory since increased EGFR is associated with increased lymph node metastasis (70) but increased CD3 expression has been associated with decreased lymph node metastasis (38). It is unclear how the presence of CD3+ T cells overrides the tumor promoting effects of EGFR.
Limitations of this study are that 1 – the work was conducted in a retrospective fashion; 2 - all cases were from a single institution; 3 – TMAs allow for only a limited area of tumor (and stroma) components for evaluation; 4 – TMA consisted of a relatively small sample size; and 5 – lack of HPV+ OSCCs in the patient cohort. Although HPV OSCCs are rare, there is evidence of HPV’s role in predicting patient prognosis (71) and there may be a benefit to including HPV+ cases in our future studies. Lastly, it is acknowledged that the expression of inhibitory checkpoints including programmed cell death-ligand 1 (PD-L1) and programmed cell death-protein 1 (PD1) regulate T-cell response (72, 73). Future studies will determine if immune checkpoint marker expression would explain some of the contradictory results observed in the present studies.
Overall, our findings suggest that the expression of CD3 was superior to CD4 and CD8 at enhancing the prognostic value of EGFR in OSCC patients. This work warrants further investigation of EGFR and CD3 as a combined prognostic biomarker profile for OSCC patients in larger patient cohorts to strengthen our findings. If successful. this work has profound implications for potential treatment options for EGFR+ patients which generally have poor clinical outcomes.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, upon reasonable request.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Review Board of the University of Iowa (IRB #201906841). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Conception and design: AS, Development of methodology: WW, AS, KG-C, AR, MB, EL, Acquisition of data: WW, KG-C, AC, Analysis and interpretation of data: WW, AS, KG-C, AR, Writing, review, and/or revision of the manuscript: WW, AS, KG-C, AR, MB, EL, Study supervision: AS, AR. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by Merit Review Award #I01 BX004829-01 from the United States (U.S.) Department of Veterans Affairs Biomedical Laboratory Research and Development Service.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to acknowledge Georgina Ofori-Amanfo and Mariah Leidinger of the Comparative Pathology Laboratory and Lisa Horning and Ellen Abusada of the Department of Pathology at the University of Iowa for their skill and expertise in the immunostaining of the OSCC tissue microarrays and Dr. Patrick Ten Eyck, the Institute for Clinical and Translational Sciences (ICTS), and support by NIH CTSA UL1TR002537 for assistance with the statistical analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.885236/full#supplementary-material
References
2. Gupta B, Johnson NW, Kumar N. Global Epidemiology of Head and Neck Cancers: A Continuing Challenge. Oncology (2016) 91(1):13–23. doi: 10.1159/000446117
3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
4. Rivera C, Oliveira AK, Costa RAP, De Rossi T, Paes Leme AF. Prognostic Biomarkers in Oral Squamous Cell Carcinoma: A Systematic Review. Oral Oncol (2017) 72:38–47. doi: 10.1016/j.oraloncology.2017.07.003
5. Argiris A, Harrington KJ, Tahara M, Schulten J, Chomette P, Ferreira Castro A, et al. Evidence-Based Treatment Options in Recurrent and/or Metastatic Squamous Cell Carcinoma of the Head and Neck. Front Oncol (2017) 7:72. doi: 10.3389/fonc.2017.00072
6. Kreppel M, Nazarli P, Grandoch A, Safi AF, Zirk M, Nickenig HJ, et al. Clinical and Histopathological Staging in Oral Squamous Cell Carcinoma - Comparison of the Prognostic Significance. Oral Oncol (2016) 60:68–73. doi: 10.1016/j.oraloncology.2016.07.004
7. Costa V, Kowalski LP, Coutinho-Camillo CM, Begnami MD, Calsavara VF, Neves JI, et al. EGFR Amplification and Expression in Oral Squamous Cell Carcinoma in Young Adults. Int J Oral Maxillofac Surg (2018) 47(7):817–23. doi: 10.1016/j.ijom.2018.01.002
8. Taghavi N, Yazdi I. Prognostic Factors of Survival Rate in Oral Squamous Cell Carcinoma: Clinical, Histologic, Genetic and Molecular Concepts. Arch Iran Med (2015) 18(5):314–9.
9. Bertics PJ, Gill GN. Self-Phosphorylation Enhances the Protein-Tyrosine Kinase Activity of the Epidermal Growth Factor Receptor. J Biol Chem (1985) 260(27):14642–7. doi: 10.1016/S0021-9258(17)38618-0
10. Bertics PJ, Weber W, Cochet C, Gill GN. Regulation of the Epidermal Growth Factor Receptor by Phosphorylation. J Cell Biochem (1985) 29(3):195–208. doi: 10.1002/jcb.240290304
11. Wee P, Wang Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers (Basel) (2017) 9(5):52. doi: 10.3390/cancers9050052
12. Romano R, Bucci C. Role of EGFR in the Nervous System. Cells (2020) 9(8):1887. doi: 10.3390/cells9081887
13. Sigismund S, Avanzato D, Lanzetti L. Emerging Functions of the EGFR in Cancer. Mol Oncol (2018) 12(1):3–20. doi: 10.1002/1878-0261.12155
14. Joseph SR, Endo-Munoz L, Gaffney DC, Saunders NA, Simpson F. Dysregulation of Epidermal Growth Factor Receptor in Actinic Keratosis and Squamous Cell Carcinoma. Curr Probl Dermatol (2015) 46:20–7. doi: 10.1159/000367959
15. Kuttan NA, Bhakthan NM. Epidermal Growth Factor Receptor (EGFR) in Oral Squamous Cell Carcinomas: Overexpression, Localization and Therapeutic Implications. Indian J Dent Res (1997) 8(1):9–18.
16. Papagerakis S, Pannone G, Zheng L, About I, Taqi N, Nguyen NP, et al. Oral Epithelial Stem Cells - Implications in Normal Development and Cancer Metastasis. Exp Cell Res (2014) 325(2):111–29. doi: 10.1016/j.yexcr.2014.04.021
17. Miyaguchi M, Olofsson J, Hellquist HB. Immunohistochemical Study of Epidermal Growth Factor Receptor in Severe Dysplasia and Carcinoma in Situ of the Vocal Cords. Acta Otolaryngol (1991) 111(1):149–52. doi: 10.3109/00016489109137366
18. Shin DM, Ro JY, Hong WK, Hittelman WN. Dysregulation of Epidermal Growth Factor Receptor Expression in Premalignant Lesions During Head and Neck Tumorigenesis. Cancer Res (1994) 54(12):3153–9.
19. Grobe A, Eichhorn W, Fraederich M, Kluwe L, Vashist Y, Wikner J, et al. Immunohistochemical and FISH Analysis of EGFR and its Prognostic Value in Patients With Oral Squamous Cell Carcinoma. J Oral Pathol Med (2014) 43(3):205–10. doi: 10.1111/jop.12111
20. Laimer K, Spizzo G, Gastl G, Obrist P, Brunhuber T, Fong D, et al. High EGFR Expression Predicts Poor Prognosis in Patients With Squamous Cell Carcinoma of the Oral Cavity and Oropharynx: A TMA-Based Immunohistochemical Analysis. Oral Oncol (2007) 43(2):193–8. doi: 10.1016/j.oraloncology.2006.02.009
21. Wheeler S, Siwak DR, Chai R, LaValle C, Seethala RR, Wang L, et al. Tumor Epidermal Growth Factor Receptor and EGFR PY1068 are Independent Prognostic Indicators for Head and Neck Squamous Cell Carcinoma. Clin Cancer Res (2012) 18(8):2278–89. doi: 10.1158/1078-0432.CCR-11-1593
22. Miyaguchi M, Olofsson J, Hellquist HB. Expression of Epidermal Growth Factor Receptor in Glottic Carcinoma and its Relation to Recurrence After Radiotherapy. Clin Otolaryngol Allied Sci (1991) 16(5):466–9. doi: 10.1111/j.1365-2273.1991.tb01041.x
23. Khaznadar SS, Khan M, Schmid E, Gebhart S, Becker ET, Krahn T, et al. EGFR Overexpression is Not Common in Patients With Head and Neck Cancer. Cell Lines are Not Representative for the Clinical Situation in This Indication. Oncotarget (2018) 9(48):28965–75. doi: 10.18632/oncotarget.25656
24. Alterio D, Marvaso G, Maffini F, Gandini S, Chiocca S, Ferrari A, et al. Role of EGFR as Prognostic Factor in Head and Neck Cancer Patients Treated With Surgery and Postoperative Radiotherapy: Proposal of a New Approach Behind the EGFR Overexpression. Med Oncol (2017) 34(6):107. doi: 10.1007/s12032-017-0965-7
25. Pedicini P, Caivano R, Jereczek-Fossa BA, Strigari L, Vischioni B, Alterio D, et al. Modelling the Correlation Between EGFr Expression and Tumour Cell Radiosensitivity, and Combined Treatments of Radiation and Monoclonal Antibody EGFr Inhibitors. Theor Biol Med Model (2012) 9:23. doi: 10.1186/1742-4682-9-23
26. Pedicini P, Nappi A, Strigari L, Jereczek-Fossa BA, Alterio D, Cremonesi M, et al. Correlation Between EGFr Expression and Accelerated Proliferation During Radiotherapy of Head and Neck Squamous Cell Carcinoma. Radiat Oncol (2012) 7:143. doi: 10.1186/1748-717X-7-143
27. Bossi P, Resteghini C, Paielli N, Licitra L, Pilotti S, Perrone F. Prognostic and Predictive Value of EGFR in Head and Neck Squamous Cell Carcinoma. Oncotarget (2016) 7(45):74362–79. doi: 10.18632/oncotarget.11413
28. Monteiro LS, Diniz-Freitas M, Garcia-Caballero T, Warnakulasuriya S, Forteza J, Fraga M. Combined Cytoplasmic and Membranous EGFR and P53 Overexpression Is a Poor Prognostic Marker in Early Stage Oral Squamous Cell Carcinoma. J Oral Pathol Med (2012) 41(7):559–67. doi: 10.1111/j.1600-0714.2012.01142.x
29. Silva SD, Alaoui-Jamali MA, Hier M, Soares FA, Graner E, Kowalski LP. Cooverexpression of ERBB1 and ERBB4 Receptors Predicts Poor Clinical Outcome in Pn+ Oral Squamous Cell Carcinoma With Extranodal Spread. Clin Exp Metastasis (2014) 31(3):307–16. doi: 10.1007/s10585-013-9629-y
30. Ulanovski D, Stern Y, Roizman P, Shpitzer T, Popovtzer A, Feinmesser R. Expression of EGFR and Cerb-B2 as Prognostic Factors in Cancer of the Tongue. Oral Oncol (2004) 40(5):532–7. doi: 10.1016/j.oraloncology.2003.11.004
31. Shiraki M, Odajima T, Ikeda T, Sasaki A, Satoh M, Yamaguchi A, et al. Combined Expression of P53, Cyclin D1 and Epidermal Growth Factor Receptor Improves Estimation of Prognosis in Curatively Resected Oral Cancer. Modern Pathol (2005) 18(11):1482–9. doi: 10.1038/modpathol.3800455
32. Monteiro L, Ricardo S, Delgado M, Garcez F, do Amaral B, Lopes C. Phosphorylated EGFR at Tyrosine 1173 Correlates With Poor Prognosis in Oral Squamous Cell Carcinomas. Oral Dis (2014) 20(2):178–85. doi: 10.1111/odi.12087
33. Monteiro LS, Diniz-Freitas M, Garcia-Caballero T, Forteza J, Fraga M. EGFR and Ki-67 Expression in Oral Squamous Cell Carcinoma Using Tissue Microarray Technology. J Oral Pathol Med (2010) 39(7):571–8. doi: 10.1111/j.1600-0714.2009.00876.x
34. Na II, Kang HJ, Cho SY, Koh JS, Lee JK, Lee BC, et al. EGFR Mutations and Human Papillomavirus in Squamous Cell Carcinoma of Tongue and Tonsil. Eur J Cancer (2007) 43(3):520–6. doi: 10.1016/j.ejca.2006.09.025
35. Nakata Y, Uzawa N, Takahashi K, Sumino J, Michikawa C, Sato H, et al. EGFR Gene Copy Number Alteration is a Better Prognostic Indicator Than Protein Overexpression in Oral Tongue Squamous Cell Carcinomas. Eur J Cancer (2011) 47(15):2364–72. doi: 10.1016/j.ejca.2011.07.006
36. Ryott M, Wangsa D, Heselmeyer-Haddad K, Lindholm J, Elmberger G, Auer G, et al. EGFR Protein Overexpression and Gene Copy Number Increases in Oral Tongue Squamous Cell Carcinoma. Eur J Cancer (2009) 45(9):1700–8. doi: 10.1016/j.ejca.2009.02.027
37. Zhou C, Wu Y, Jiang L, Li Z, Diao P, Wang D, et al. Density and Location of CD3(+) and CD8(+) Tumor-Infiltrating Lymphocytes Correlate With Prognosis of Oral Squamous Cell Carcinoma. J Oral Pathol Med (2018) 47(4):359–67. doi: 10.1111/jop.12698
38. Mukherjee G, Bag S, Chakraborty P, Dey D, Roy S, Jain P, et al. Density of CD3+ and CD8+ Cells in Gingivo-Buccal Oral Squamous Cell Carcinoma is Associated With Lymph Node Metastases and Survival. PloS One (2020) 15(11):e0242058. doi: 10.1371/journal.pone.0242058
39. Spector ME, Bellile E, Amlani L, Zarins K, Smith J, Brenner JC, et al. Prognostic Value of Tumor-Infiltrating Lymphocytes in Head and Neck Squamous Cell Carcinoma. JAMA Otolaryngology–Head Neck Surgery (2019) 145(11):1012–9. doi: 10.1001/jamaoto.2019.2427
40. Watanabe Y, Katou F, Ohtani H, Nakayama T, Yoshie O, Hashimoto K. Tumor-Infiltrating Lymphocytes, Particularly the Balance Between CD8(+) T Cells and CCR4(+) Regulatory T Cells, Affect the Survival of Patients With Oral Squamous Cell Carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod (2010) 109(5):744–52. doi: 10.1016/j.tripleo.2009.12.015
41. Ni YH, Zhang XX, Lu ZY, Huang XF, Wang ZY, Yang Y, et al. Tumor-Infiltrating CD1a(+) DCs and CD8(+)/FoxP3(+) Ratios Served as Predictors for Clinical Outcomes in Tongue Squamous Cell Carcinoma Patients. Pathol Oncol Res (2020) 26(3):1687–95. doi: 10.1007/s12253-019-00701-5
42. Diao P, Jiang Y, Li Y, Wu X, Li J, Zhou C, et al. Immune Landscape and Subtypes in Primary Resectable Oral Squamous Cell Carcinoma: Prognostic Significance and Predictive of Therapeutic Response. J Immunother Cancer (2021) 9(6):e002434. doi: 10.1136/jitc-2021-002434
43. Quan H, Shan Z, Liu Z, Liu S, Yang L, Fang X, et al. The Repertoire of Tumor-Infiltrating Lymphocytes Within the Microenvironment of Oral Squamous Cell Carcinoma Reveals Immune Dysfunction. Cancer Immunol Immunother (2020) 69(3):465–76. doi: 10.1007/s00262-020-02479-x
44. Troiano G, Rubini C, Togni L, Caponio VCA, Zhurakivska K, Santarelli A, et al. The Immune Phenotype of Tongue Squamous Cell Carcinoma Predicts Early Relapse and Poor Prognosis. Cancer Med (2020) 9(22):8333–44. doi: 10.1002/cam4.3440
45. Lewis CE, Pollard JW. Distinct Role of Macrophages in Different Tumor Microenvironments. Cancer Res (2006) 66(2):605–12. doi: 10.1158/0008-5472.CAN-05-4005
46. Fukaya T, Takagi H, Taya H, Sato K. DCs in Immune Tolerance in Steady-State Conditions. Methods Mol Biol (2011) 677:113–26. doi: 10.1007/978-1-60761-869-0_8
47. Facciabene A, Motz GT, Coukos G. T-Regulatory Cells: Key Players in Tumor Immune Escape and Angiogenesis. Cancer Res (2012) 72(9):2162–71. doi: 10.1158/0008-5472.CAN-11-3687
48. Malek E, de Lima M, Letterio JJ, Kim BG, Finke JH, Driscoll JJ, et al. Myeloid-Derived Suppressor Cells: The Green Light for Myeloma Immune Escape. Blood Rev (2016) 30(5):341–8. doi: 10.1016/j.blre.2016.04.002
49. Gonzalez H, Hagerling C, Werb Z. Roles of the Immune System in Cancer: From Tumor Initiation to Metastatic Progression. Genes Dev (2018) 32(19-20):1267–84. doi: 10.1101/gad.314617.118
50. Lequerica-Fernandez P, Suarez-Canto J, Rodriguez-Santamarta T, Rodrigo JP, Suarez-Sanchez FJ, Blanco-Lorenzo V, et al. Prognostic Relevance of CD4(+), CD8(+) and FOXP3(+) TILs in Oral Squamous Cell Carcinoma and Correlations With PD-L1 and Cancer Stem Cell Markers. Biomedicines (2021) 9(6):653. doi: 10.3390/biomedicines9060653
51. Shimizu S, Hiratsuka H, Koike K, Tsuchihashi K, Sonoda T, Ogi K, et al. Tumor-Infiltrating CD8(+) T-Cell Density is an Independent Prognostic Marker for Oral Squamous Cell Carcinoma. Cancer Med (2019) 8(1):80–93. doi: 10.1002/cam4.1889
52. Wu J, Zhang T, Xiong H, Zeng L, Wang Z, Peng Y, et al. Tumor-Infiltrating CD4(+) Central Memory T Cells Correlated With Favorable Prognosis in Oral Squamous Cell Carcinoma. J Inflammation Res (2022) 15:141–52. doi: 10.2147/JIR.S343432
53. Balermpas P, Michel Y, Wagenblast J, Seitz O, Weiss C, Rodel F, et al. Tumour-Infiltrating Lymphocytes Predict Response to Definitive Chemoradiotherapy in Head and Neck Cancer. Br J Cancer (2014) 110(2):501–9. doi: 10.1038/bjc.2013.640
54. Nguyen N, Bellile E, Thomas D, McHugh J, Rozek L, Virani S, et al. Tumor Infiltrating Lymphocytes and Survival in Patients With Head and Neck Squamous Cell Carcinoma. Head Neck (2016) 38(7):1074–84. doi: 10.1002/hed.24406
55. Nordfors C, Grun N, Tertipis N, Ahrlund-Richter A, Haeggblom L, Sivars L, et al. CD8+ and CD4+ Tumour Infiltrating Lymphocytes in Relation to Human Papillomavirus Status and Clinical Outcome in Tonsillar and Base of Tongue Squamous Cell Carcinoma. Eur J Cancer (2013) 49(11):2522–30. doi: 10.1016/j.ejca.2013.03.019
56. Gamboa-Dominguez A, Dominguez-Fonseca C, Quintanilla-Martinez L, Reyes-Gutierrez E, Green D, Angeles-Angeles A, et al. Epidermal Growth Factor Receptor Expression Correlates With Poor Survival in Gastric Adenocarcinoma From Mexican Patients: A Multivariate Analysis Using a Standardized Immunohistochemical Detection System. Modern Pathol (2004) 17(5):579–87. doi: 10.1038/modpathol.3800085
57. Bourova-Flin E, Derakhshan S, Goudarzi A, Wang T, Vitte A-L, Chuffart F, et al. The Combined Detection of Amphiregulin, Cyclin A1 and DDX20/Gemin3 Expression Predicts Aggressive Forms of Oral Squamous Cell Carcinoma. Br J Cancer (2021) 125(8):1122–34. doi: 10.1038/s41416-021-01491-x
58. Rajan A, Gibson-Corley KN, Choi AB, Ofori-Amanfo GK, Ten Eyck P, Espinosa-Cotton M, et al. Impact of Nuclear Interleukin-1 Alpha and EGFR Expression on Recurrence and Survival Outcomes in Oral Squamous Cell Carcinomas. J Oncol (2019) 2019:5859680. doi: 10.1155/2019/5859680
59. Polanska H, Raudenska M, Hudcova K, Gumulec J, Svobodova M, Heger Z, et al. Evaluation of EGFR as a Prognostic and Diagnostic Marker for Head and Neck Squamous Cell Carcinoma Patients. Oncol Lett (2016) 12(3):2127–32. doi: 10.3892/ol.2016.4896
60. Ma P, He Q, Li W, Li X, Han H, Jin M, et al. Anti-CD3 X EGFR Bispecific Antibody Redirects Cytokine-Induced Killer Cells to Glioblastoma In Vitro and In Vivo. Oncol Rep (2015) 34(5):2567–75. doi: 10.3892/or.2015.4233
61. Chai AWY, Lim KP, Cheong SC. Translational Genomics and Recent Advances in Oral Squamous Cell Carcinoma. Semin Cancer Biol (2020) 61:71–83. doi: 10.1016/j.semcancer.2019.09.011
62. Sales de Sá R, Miranda Galvis M, Mariz BALA, Leite AA, Schultz L, Almeida OP, et al. Increased Tumor Immune Microenvironment CD3+ and CD20+ Lymphocytes Predict a Better Prognosis in Oral Tongue Squamous Cell Carcinoma. Front Cell Dev Biol (2021) 8:622161. doi: 10.3389/fcell.2020.622161
63. Badoual C, Hans S, Rodriguez J, Peyrard S, Klein C, Agueznay Nel H, et al. Prognostic Value of Tumor-Infiltrating CD4+ T-Cell Subpopulations in Head and Neck Cancers. Clin Cancer Res (2006) 12(2):465–72. doi: 10.1158/1078-0432.CCR-05-1886
64. Wu RQ, Zhang DF, Tu E, Chen QM, Chen W. The Mucosal Immune System in the Oral Cavity-an Orchestra of T Cell Diversity. Int J Oral Sci (2014) 6(3):125–32. doi: 10.1038/ijos.2014.48
65. Chatzopoulos K, Sotiriou S, Collins AR, Kartsidis P, Schmitt AC, Chen X, et al. Transcriptomic and Immunophenotypic Characterization of Tumor Immune Microenvironment in Squamous Cell Carcinoma of the Oral Tongue. Head Neck Pathol (2021) 15(2):509–22. doi: 10.1007/s12105-020-01229-w
66. Heikkinen I, Bello IO, Wahab A, Hagstrom J, Haglund C, Coletta RD, et al. Assessment of Tumor-Infiltrating Lymphocytes Predicts the Behavior of Early-Stage Oral Tongue Cancer. Am J Surg Pathol (2019) 43(10):1392–6. doi: 10.1097/PAS.0000000000001323
67. Lee HJ, Kim JY, Park IA, Song IH, Yu JH, Ahn JH, et al. Prognostic Significance of Tumor-Infiltrating Lymphocytes and the Tertiary Lymphoid Structures in HER2-Positive Breast Cancer Treated With Adjuvant Trastuzumab. Am J Clin Pathol (2015) 144(2):278–88. doi: 10.1309/AJCPIXUYDVZ0RZ3G
68. Meehan K, Leslie C, Lucas M, Jacques A, Mirzai B, Lim J, et al. Characterization of the Immune Profile of Oral Tongue Squamous Cell Carcinomas With Advancing Disease. Cancer Med (2020) 9(13):4791–807. doi: 10.1002/cam4.3106
69. Schuler PJ, Boeckers P, Engers R, Boelke E, Bas M, Greve J, et al. EGFR-Specific T Cell Frequencies Correlate With EGFR Expression in Head and Neck Squamous Cell Carcinoma. J Transl Med (2011) 9:168. doi: 10.1186/1479-5876-9-168
70. Lu Y, Jingyan G, Baorong S, Peng J, Xu Y, Cai S. Expression of EGFR, Her2 Predict Lymph Node Metastasis (LNM)-Associated Metastasis in Colorectal Cancer. Cancer biomark (2012) 11(5):219–26. doi: 10.3233/CBM-2012-00282
71. Christianto S, Li KY, Huang TH, Su YX. The Prognostic Value of Human Papilloma Virus Infection in Oral Cavity Squamous Cell Carcinoma: A Meta-Analysis. Laryngoscope (2021). doi: 10.1002/lary.29996
72. de Vicente JC, Rodríguez-Santamarta T, Rodrigo JP, Blanco-Lorenzo V, Allonca E, García-Pedrero JM. PD-L1 Expression in Tumor Cells Is an Independent Unfavorable Prognostic Factor in Oral Squamous Cell Carcinoma. Cancer Epidemiol Biomarkers Prev (2019) 28(3):546–54. doi: 10.1158/1055-9965.EPI-18-0779
Keywords: OSCC, EGFR, CD3, TIL, microarray, biomarker
Citation: Wongpattaraworakul W, Gibson-Corley KN, Choi A, Buchakjian MR, Lanzel EA, Rajan KD A and Simons AL (2022) Prognostic Role of Combined EGFR and Tumor-Infiltrating Lymphocytes in Oral Squamous Cell Carcinoma. Front. Oncol. 12:885236. doi: 10.3389/fonc.2022.885236
Received: 27 February 2022; Accepted: 22 June 2022;
Published: 25 July 2022.
Edited by:
Steve Oghumu, The Ohio State University, United StatesReviewed by:
Marco Mascitti, Marche Polytechnic University, ItalyTarig Osman, University of Bergen, Norway
Copyright © 2022 Wongpattaraworakul, Gibson-Corley, Choi, Buchakjian, Lanzel, Rajan KD and Simons. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrean L. Simons, andrean-simons@uiowa.edu
 Wattawan Wongpattaraworakul
Wattawan Wongpattaraworakul Katherine N. Gibson-Corley
Katherine N. Gibson-Corley Allen Choi2
Allen Choi2 Marisa R. Buchakjian
Marisa R. Buchakjian Anand Rajan KD
Anand Rajan KD Andrean L. Simons
Andrean L. Simons