Abstract
Class III phosphatidylinositol 3-kinase (PI3-kinase) regulates multiple membrane trafficking. In yeast, two distinct PI3-kinase complexes are known: complex I (Vps34, Vps15, Vps30/Atg6, and Atg14) is involved in autophagy, and complex II (Vps34, Vps15, Vps30/Atg6, and Vps38) functions in the vacuolar protein sorting pathway. Atg14 and Vps38 are important in inducing both complexes to exert distinct functions. In mammals, the counterparts of Vps34, Vps15, and Vps30/Atg6 have been identified as Vps34, p150, and Beclin 1, respectively. However, orthologues of Atg14 and Vps38 remain unknown. We identified putative mammalian homologues of Atg14 and Vps38. The Vps38 candidate is identical to UV irradiation resistance-associated gene (UVRAG), which has been reported as a Beclin 1-interacting protein. Although both human Atg14 and UVRAG interact with Beclin 1 and Vps34, Atg14, and UVRAG are not present in the same complex. Although Atg14 is present on autophagic isolation membranes, UVRAG primarily associates with Rab9-positive endosomes. Silencing of human Atg14 in HeLa cells suppresses autophagosome formation. The coiled-coil region of Atg14 required for binding with Vps34 and Beclin 1 is essential for autophagy. These results suggest that mammalian cells have at least two distinct class III PI3-kinase complexes, which may function in different membrane trafficking pathways.
INTRODUCTION
Phosphatidylinositol 3-kinases (PI3-kinases) are divided into three classes based on structure of the catalytic subunits. Class I, and probably class II PI3-kinases play important roles in signal transduction after various external stimuli (Vanhaesebroeck et al., 2001). In contrast, class III PI3-kinase is involved in intracellular membrane trafficking (Lindmo and Stenmark, 2006). Yeast cells have only class III PI3-kinase, Vps34 (Schu et al., 1993). Vps34 forms at least two distinct PI3-kinase complexes: type 1 complex consisting of Vps34, Vps15, Atg6/Vps30 and Atg14, and type 2 complex containing Vps34, Vps15, Atg6/Vps30, and Vps38. Type 1 and 2 complexes function in autophagy and the vacuolar protein sorting pathway, respectively (Kihara et al., 2001b). Although a myristoylated protein kinase Vps15 (Stack et al., 1993) and Atg6/Vps30 (Kametaka et al., 1998) are shared subunits, Atg14 and Vps38 are subunits specific to each complex.
Macroautophagy, hereafter referred to simply as autophagy, is a bulk degradation process occurring in cells and is conserved in all eukaryotes (Cuervo, 2004; Levine and Klionsky, 2004; Klionsky, 2007; Mizushima, 2007; Mizushima et al., 2008). A portion of the cytoplasm is sequestered by an autophagosome, which then fuses with a lysosome (or vacuole in yeast and plants) to degrade the materials inside. Yeast genetic analyses have identified at least 31 genes (ATG genes) involved in autophagy and its related pathways (Klionsky, 2005; Suzuki and Ohsumi, 2007). More than half of the ATG gene products (Atg1-10, 12–14, 16–18, 29, and 31) are required for autophagosome formation, and they are therefore known as AP-Atg proteins (Kabeya et al., 2007; Suzuki and Ohsumi, 2007). Almost all AP-Atg proteins are localized to a perivacuolar structure called the preautophagosomal structure (PAS), where autophagosomes are generated (Kim et al., 2001; Suzuki et al., 2001). Atg14 localizes in the PAS and vacuolar membrane, and it mediates PAS targeting of Vps34 and Atg6/Vps30 (Obara et al., 2006). This type 1 PI3-kinase complex is important for proper PAS targeting of other Atg proteins such as Atg8, the Atg12-Atg5 · Atg16 complex, Atg2 and Atg18 (Suzuki et al., 2007).
The vacuolar protein sorting pathway is completely distinct from autophagy. Vacuolar proteins are sorted at the trans-Golgi network (TGN) and delivered to the vacuole. Vps38 is present on endosomes and vacuolar membranes, and it is required for endosome targeting of Vps30 and Vps34 (Obara et al., 2006). Therefore, Atg14 and Vps38 mediate formation of type 1 and type 2 PI3-kinase complexes on their proper membranes (Obara et al., 2006).
The downstream effectors of this PI3-kinase complex have not been completely elucidated. Atg18 (Dove et al., 2004; Stromhaug et al., 2004; Obara et al., 2008b), Atg20 (Nice et al., 2002), Atg21 (Stromhaug et al., 2004), Atg24/Snx4 (Nice et al., 2002; Hettema et al., 2003), and Atg27 (Wurmser and Emr, 2002) have been identified to bind phosphatidylinositol 3-phosphate [PI(3)P]. Although Atg18 is required for autophagy, others are required only for the cytoplasm-to-vacuole targeting pathway (Klionsky, 2005; Obara et al., 2008b). The downstream effectors of the type 2 complex are thought to be Vps5 and Vps17, both of which possess the Phox homology (PX) domain (Burda et al., 2002). Vps5 and Vps17 are included in the retromer complex together with Vps26, Vps29, and Vps35.
The mammalian class III PI3-kinase has also been suggested to be important for various membrane trafficking pathways such as the Golgi-to-lysosome pathway (Brown et al., 1995; Davidson, 1995), internal vesicle formation in late endosomes (Futter et al., 2001; Johnson et al., 2006), and autophagy (Petiot et al., 2000; Kihara et al., 2001a). It is well known that mammalian cells have the class III PI3-kinase complex consisting of Vps34 and its regulatory subunit called p150, which is a Vps15 counterpart (Vanhaesebroeck et al., 2001; Lindmo and Stenmark, 2006). Beclin 1, which is the mammalian homologue of Atg6/Vps30, is also contained in this protein complex (Liang et al., 1999; Kihara et al., 2001a). Beclin 1 was originally identified as an interacting partner of an antiapoptotic protein Bcl-2 (Liang et al., 1998). Bcl-2 negatively regulates autophagy through binding with Beclin 1 (Pattingre et al., 2005; Maiuri et al., 2007; Wei et al., 2008). Moreover, Beclin 1 is also known as a tumor suppressor (Liang et al., 1999; Qu et al., 2003; Yue et al., 2003). Recently, Beclin 1 was found to interact with two additional molecules, UV irradiation resistance-associated gene (UVRAG) (Liang et al., 2006) and a WD-40 domain-containing protein named Ambra1 (Maria Fimia et al., 2007). Both UVRAG and Ambra1 are present in the human Vps34 complex and seem to be positive regulators of autophagy.
Although it has been well established that the Beclin 1 complex is required for autophagy, the involvement of Atg6/Vps30/Beclin 1 in other membrane trafficking pathways remains controversial in species other than yeast. Expression of plant Atg6 in yeast atg6-deficient cells restores both autophagy and vacuolar protein sorting abilities (Fujiki et al., 2007). Furthermore, Arabidopsis atg6 mutant shows defects in autophagy and pollen germination, but the latter was not observed in other autophagy mutants, suggesting that Atg6 has some autophagy-independent function (Fujiki et al., 2007). In Caenorhabditis elegans, BEC-1/Atg6 was shown to be required for both autophagy and endocytosis (Takacs-Vellai et al., 2005). In contrast, mammalian Beclin 1 has been shown to restore autophagy but not vacuolar protein sorting in yeast atg6 mutant (Liang et al., 1999). In addition, RNA interference (RNAi)-mediated suppression of Beclin 1 impairs autophagy but does not affect the endocytic sorting of the epidermal growth factor receptor and cathepsin D transport from TGN to lysosomes (Zeng et al., 2006). Cathepsin D maturation also seems normal in Beclin 1-deficient MCF7 cells, although autophagy is suppressed in these cells (Furuya et al., 2005).
These results imply that Beclin 1 could be involved solely in autophagy. However, considering that Beclin 1 knockout mice die at approximately embryonic day 7.5 (Yue et al., 2003), whereas Atg5 and Atg7 knockout mice can survive until birth (Kuma et al., 2004; Komatsu et al., 2005), Beclin 1 might have more complicated functions other than autophagy. It is therefore reasonable to assume that mammalian cells, like yeast cells, have several distinct PI3-kinase complexes. However, although Beclin 1 was discovered ∼10 years ago, homologues of Atg14 and Vps38 have remained unknown.
Here, we report identification of human Atg14 and Vps38 candidates. Human Atg14 and Vps38 form distinct PI3-kinase complexes showing different intracellular localization. These results suggest that Vps34–Beclin 1 complexes may have at least two different functions as in yeast cells.
MATERIALS AND METHODS
Cell Culture and Transfection
HeLa and human embryonic kidney (HEK)293T cells were cultured in DMEM supplemented with 10% fatal bovine serum (FBS), 50 μg/ml penicillin, and streptomycin in a 5% CO2 incubator. NIN3T3 cells were maintained in DMEM containing 10% bovine calf serum and the antibiotics. For starvation, cells were washed with phosphate-buffered saline (PBS) and incubated in amino acid-free DMEM without FBS. FuGENE 6 reagent (Roche Diagnostics, Tokyo, Japan) or Lipofectamine 2000 reagent (Invitrogen, Tokyo, Japan) were used for transfection. Stable cell lines were generated using a retroviral expression system (Hara et al., 2008). E64d and pepstatin were purchased from Peptide Institute (Osaka, Japan), and chloroquine and wortmannin were purchased from Sigma-Aldrich (St. Louis, MO).
Plasmids
cDNAs encoding human Atg14 (AK131251/FLJ 16178, obtained from the Department of Biotechnology, National Institute of Technology and Evaluation, Tokyo, Japan) and human UVRAG (IMAGE 618651) were amplified by polymerase chain reaction (PCR) and subcloned into p3xFLAG CMV10 (Sigma-Aldrich) or pCI-neo (Promega, Tokyo, Japan) together with hemagglutinin (HA) or Myc epitope tag. pCMV5 HA-Beclin 1, pcDNA3 FLAG-Vps34, and pcDNA3 MEF-p150 plasmids were kindly provided by Dr. Tamotsu Yoshimori (Osaka University, Osaka, Japan). FLAG-Rab5, FLAG-Rab7, and FLAG-Rab9 plasmids were kindly provided by Dr. Mitsunori Fukuda (Tohoku University, Sendai, Japan). Rab9 was subcloned into pCI-neo together with the HA epitope tag. For retroviral expression, cDNAs encoding human Atg14, UVRAG, and Vps34 were subcloned into pMXs-puro or pMXs-IP vector (provided by T. Kitamura, The University of Tokyo, Tokyo, Japan). A small interference RNA (siRNA)-resistant Atg14 silent mutant was created by substituting four nucleotides in the Atg14 siRNA-targeting region (A582T, A585T, C588A, and T591A).
Antibodies
To generate antibodies against human Atg14 and UVRAG, human Atg14 cDNA (amino acid residues 270–404) and human UVRAG cDNA (amino acid residues 137-419) were subcloned into pGEX-6p-1 (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Resulting glutathione transferase (GST)-fused Atg14 and UVRAG were used to immunize rabbits. Antisera were affinity-purified. Mouse monoclonal anti-HA (16B12) and anti-Myc (9E10) antibodies, and rabbit polyclonal anti-HA antibody, were purchased from Covance Research Products (Princeton, NJ). Mouse monoclonal anti-FLAG antibody (M2), anti-β-actin, and anti-Atg5 antibody were purchased from Sigma-Aldrich. Rat monoclonal anti-green fluorescent protein (GFP) antibody (GF090R) was purchased from Nacalai Tesque (Kyoto, Japan) and used for immunocytochemistry. Rabbit polyclonal anti-GFP antibody was purchased from Invitrogen and used for immunoprecipitation and immunoblotting. Guinea pig polyclonal anti-p62 antibody was purchased from Progen (Heidelberg, Germany). The polyclonal antibodies against microtubule-associated protein light chain 3 (LC3) (SK2-6) (Kabeya et al., 2000), Vps34, Beclin 1 (Kihara et al., 2001a), and Atg16L1 (Mizushima et al., 2003) were described previously.
Immunoprecipitation and Immunoblotting
Cell lysates were prepared in a lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100, 1 mM phenylmethanesulfonyl fluoride, 1 mM Na3VO4, and protease inhibitor cocktail [Complete EDTA-free protease inhibitor; Roche Diagnostics]). The lysates were clarified by centrifugation at 14,000 rpm for 15 min and were subjected to immunoprecipitation by using specific antibodies in combination with protein G-Sepharose (GE Healthcare). Precipitated immunocomplexes were washed 6 times in a wash buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100) and boiled in sample buffer. Samples were subsequently separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to Immobilon-P polyvinylidene diflouride membranes (Millipore, Billerica, MA). Immunoblot analysis was performed with the indicated antibodies and visualized with SuperSignal West Pico Chemiluminescent substrate (Pierce Chemical, Rockford, IL).
Immunocytochemistry
Cells grown on coverslips were washed with PBS and fixed in 4% paraformaldehyde in PBS for 10 min at room temperature. Fixed cells were permeabilized with 50 μg/ml digitonin in PBS for 15 min, blocked with 3% bovine serum albumin in PBS and incubated with primary antibody for 1 h. After washing, cells were incubated with AlexaFluor 488-conjugated goat anti-rat immunoglobulin G (IgG) or AlexaFluor 564-conjugated goat anti-rabbit or anti-mouse IgG secondary antibody (Invitrogen) for 1 h and examined under a fluorescence microscope (IX81; Olympus, Tokyo, Japan) equipped with a charge-coupled device camera (ORCA ER; Hamamatsu Photonics, Hamamatsu, Japan). Quantification of GFP-LC3 dots was performed using the Top Hat algorithm of MetaMorph version 7.0 (Molecular Devices, Tokyo, Japan) as described previously (Kuma et al., 2004).
RNAi
Stealth RNAi oligonucleotides were used for siRNA experiments (Invitrogen). The sequences used were as follows: human Atg14-1-siRNA sense, 5′-GGCAAAUCUUCGACGAUCCCAUAUA-3′ and antisense, 5′-UAUAUGGGAUCGUCGAAGAUUUGCC-3′; human Atg14-2-siRNA sense, 5′-GAUCACAACGGAGACACCAGCAUUA-3′ and antisense, 5′-UAAUGCUGGUGUCUCCGUUGUGAUC-3′; human UVRAG-siRNA sense, 5′-CAUCAGCUCCUUGAUACCUACUUUA-3′ and antisense, 5′-UAAAGUAGGUAUCAAGGAGCUGAUG-3′; uman Beclin 1-siRNA sense, 5′-CCACUCUGUGAGGAAUGCACAGAUA-3′ and antisense, 5′-UAUCUGUGCAUUCCUCACAGAGUGG-3′; and human Vps34-siRNA sense, 5′-CCCAUGAGAUGUACUUGAACGUAAU-3′ and antisense, 5′-AUUACGUUCAAGUACAUCUCAUGGG-3′.
For a negative control, Medium GC duplex of stealth RNAi Negative Control Duplexes (Invitrogen) was used. The Stealth RNAi oligonucleotides were transfected into HeLa cells by using Lipofectamine RNAi MAX according to the manufacturer's protocols. After 2 d, the cells were again transfected with the same siRNA and cultured for additional 3 d before analysis, unless otherwise specified.
Reverse Transcriptase (RT)-PCR
Total RNA was extracted from mouse tissues using Isogen (Nippon Gene, Toyama, Japan) according to the manufacturer's instruction. Trace contamination of DNA was removed by DNase digestion (Promega). cDNA was synthesized from 2 μg of total RNA by using ReverTra Ace (Toyobo Engineering, Osaka, Japan). PCR amplification was performed using the following primers: mouse Atg14 forward, 5′-CATAACAACCCCGCCTACAC-3′ and reverse, 5′-TGCGTTCAGTTTCCTCACTG-3′; mouse UVRAG forward, 5′-GCAGAAGGACTCCCTGAGTG-3′ and reverse, 5′-GGAAGTCCTCGGAATTAGGC-3′; and mouse β-actin forward, 5′-CTGGGTATGGAATCCTGTGG-3′; reverse, 5′-GTACTTGCGCTCAGGAGGAG-3′.
Real-Time PCR
Real-time PCR was performed on a Thermal Cycler Dice (Takara, Kyoto, Japan) by using SYBR premix EX Taq (Takara). The primer sets used were as follows: human Vps34 forward, 5′-AAGCAGTGCCTGTAGGAGGA-3′ and reverse, 5′-TGTCGATGAGCTTTGGTGAG-3′; human Beclin 1 forward, 5′-TGTCACCATCCAGGAACTCA-3′ and reverse, 5′-CTGTTGGCACTTTCTGTGGA-3′; human Atg14 forward, 5′-ATGAGCGTCTGGCAAATCTT-3′ and reverse, 5′-CCCATCGTCCTGAGAGGTAA-3′; human UVRAG forward, 5′-CTGTTTGGATGGGCTGAAAT-3′ and reverse, 5′-TGCGAACACAGTTCTGATCC-3′; and human β-actin forward, 5′-ATTGCCGACAGGATGCAGAA-3′ and reverse, 5′-ACATCTGCTGGAAGGTGGACAG-3′. Amplicon expression in each sample was normalized to its β-actin mRNA content.
Electron Microscopy
Conventional electron microscopy was performed as described previously (Hara et al., 2008). For morphometric analysis of autophagosomes, at least 10 cells of each sample were analyzed using MetaMorph image analysis software, version 6.2 (MDS Analytical Technologies, Sunnyvale, CA).
Yeast Studies
Total cell lysates were prepared and subjected to immunoblot analysis as described previously (Mizushima et al., 1999). Separation of extracellular and intracellular forms of carboxypeptidase Y (CPY) was performed essentially as described previously (Gabriely et al., 2007). Five OD600 U of mid-log phase yeast cells were incubated in 500 μl of YPD medium for 1 h at 30°C. Then, 5 μl of 1 M NaN3 was added, and the cell cultures were cooled on ice for 10 min. Culture samples were centrifuged to separate the cells (containing the intracellular fraction) from the medium (containing the extracellular fraction). The culture medium was precipitated with 10% trichloroacetic acid and 10% Triton X-100 for 10 min at 4°C and centrifuged at 15,000 rpm for 30 min. Samples were washed with chilled acetone and resuspended in 200 μl of SDS sample buffer. The yeast Atg14 and Vps38 expression plasmids, Ape1 and CPY antibodies, and Δapg14 and Δvps38 strains were generous gifts from Drs. Yoshinori Ohsumi and Keisuke Obara (National Institute for Basic Biology, Okazaki, Japan).
RESULTS
Identification of Mammalian Homologues of Atg14 and Vps38 (UVRAG)
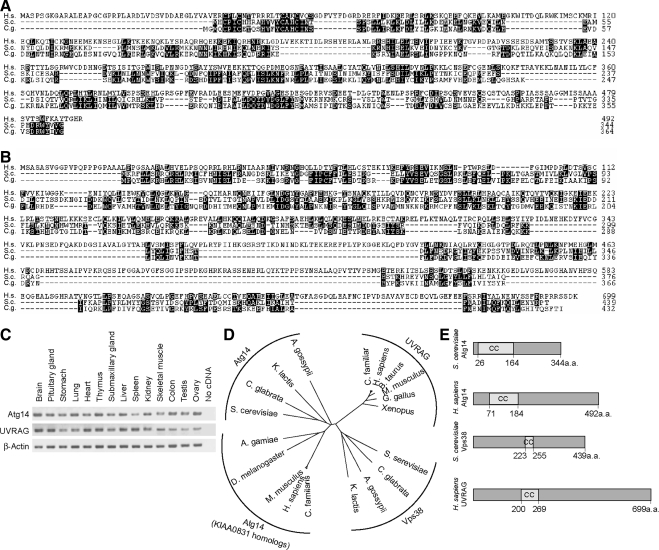
Mammalian homologues of Atg14 and Vps38 have not been successfully identified by simple BLAST database search. We therefore performed a position specific iterative (PSI)-BLAST search of the National Center for Biotechnology Information (NCBI) database by using Saccharomyces cerevisiae Atg14 (NP_009686) and Candida glabrata Atg14 (XP_445209) (Altschul et al., 1997). PSI-BLAST is more sensitive to weak but biologically relevant sequence similarities than conventional BLAST, because we can create a position-specific score matrix for each query protein sequence (Jones and Swindells, 2002). After two or three iterated profiling, a putative mammalian homologue, KIAA0831, was identified in both human and mouse databases. Human KIAA0831 (NP_055739) is a 492-amino acid protein with no obvious sequence motifs except the coiled-coil domain, and it has weak homology to S. cerevisiae Atg14 (13.1% identity and 37.2% similarity) (Figure 1A). Likewise, we performed a similar PSI-BLAST search by using S. cerevisiae Vps38 (NP_013464) and C. glabrata Vps38 (XP_445092), and identified UVRAG as a putative Vps38 homologue. UVRAG shows 10.2% identity and 31.9% similarity to yeast Vps38 (Figure 1B). Both KIAA0831 and UVRAG transcripts were widely expressed in mouse tissues (Figure 1C).
Figure 1.
Identification of mammalian counterparts of Atg14 and Vps38. (A) Amino acid alignment of Homo sapiens Atg14 (KIAA0831; National Center for Biotechnology Information [NCBI]accession no. NP_055739) with Atg14 of S. cerevisiae (accession no. NP_009686) and C. glabrata (accession no. XP_445209). The alignment was generated using CLUSTAL W. Identical residues are indicated with filled boxes. (B) Amino acid alignment of H. sapiens UVRAG (NCBI accession no. NP_003360) with Vps38 of S. cerevisiae (accession no. NP_013464) and C. glabrata (accession no. XP_445209). (C) KIAA0831 (Atg14) and UVRAG are ubiquitously expressed in mouse tissues. Total RNA from mouse tissues was reverse-transcribed into cDNA and then subjected to PCR amplification with indicated primers. (D) Phylogenetic analysis of Atg14, KIAA0831, UVRAG, and Vps38 homologues. The unrooted phylogenetic tree was constructed using CLUSTAL W based on the amino acid sequences of these homologues. Distance matrix-based trees were constructed by the NJ method (Saitou and Nei, 1987). (E) Structural comparison of S. cerevisiae Atg14, H. sapiens Atg14 (KIAA0831), S. cerevisiae Vps38, and H. sapiens UVRAG. The coiled-coil domain is predicted by the algorithm of Lupas et al. (1991).
KIAA0831 homologues were widely found in species where the Atg14 homologue has not been identified. Similarly, UVRAG homologues were found in many species in which Vps38 has been unknown. We then constructed an unrooted neighbor-joining (NJ) tree of these proteins (Figure 1D). According to the tree topology, the aligned sequences were classified into four groups: the Atg14, Vps38, KIAA0831, and UVRAG homologues. The Atg14 homologues were closely related to the KIAA0831 homologues, but not to the UVRAG homologues. The UVRAG homologues were located between the Atg14 and Vps38 homologues, but they seemed to be closer to the Vps38 homologues. Both human KIAA0831 and yeast Atg14 have a coiled-coil domain in their N-terminal region, whereas UVRAG and yeast Vps38 possess the coiled-coil domain in their center region (Figure 1E). Based on these findings together with the results described below, we named KIAA0831 as human Atg14. Our sequence analysis also suggests that UVRAG could be the functional counterpart of Vps38.
Human Atg14 Interacts with Beclin 1 and Vps34 but Not with UVRAG
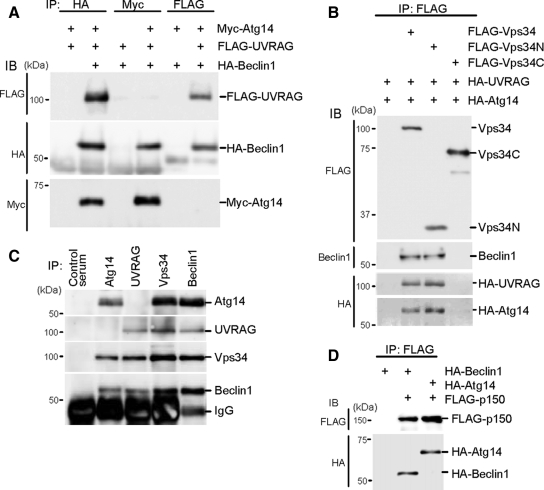
We first analyzed complex formation of human Atg14, Beclin 1, Vps34, and UVRAG. Atg14, Beclin 1, and UVRAG were N-terminally fused to the Myc-, HA-, and FLAG-tags, respectively, and transiently coexpressed in HEK293T cells. When the cell lysate was precipitated with HA-Beclin 1, both Atg14 and UVRAG were detected in the precipitates (Figure 2A). However, if we precipitated the lysate with Myc-Atg14, only Beclin 1, but not UVRAG, was coprecipitated. Likewise, Atg14 was not coprecipitated with UVRAG, whereas Beclin 1 was precipitated. These findings suggest that Atg14 and UVRAG form distinct Beclin 1 complexes. These interactions were also observed in HeLa cells and NIH3T3 cells (Supplemental Figure 1).
Figure 2.
Atg14 and UVRAG form two distinct complexes with Beclin 1-Vps34. (A) Human Atg14 and UVRAG are included in different Beclin 1 complexes. HEK293T cells were cotransfected with FLAG-UVRAG, HA-Beclin 1, and Myc-Atg14, and cell lysates were subjected to immunoprecipitation using antibodies against HA, Myc, and FLAG. The resulting precipitates were examined by immunoblot analysis with the indicated antibodies. (B) Atg14 and UVRAG interacts with the N-terminal C2 domain of Vps34. HEK293T cells were cotransfected with HA-UVRAG, HA-Atg14, and either FLAG-Vps34 (full-length), FLAG-Vps34N (containing C2 domain, 1-282 amino acid residues), or FLAG-Vps34C (283-887 amino acid residues) and analyzed as described in A. (C) Endogenous Atg14 and UVRAG form protein complexes with Vps34 and Beclin 1 but not with each other. Lysates of HEK293T cells were used for immunoprecipitation with indicated antiserum. (D) Atg14 forms a protein complex with p150. Lysates of HEK293T cells transiently expressing FLAG-p150 and either HA-Beclin 1 or HA-Atg14 were analyzed by immunoprecipitation and immunoblotting.
Vps34 contains three distinct domains: the N-terminal C2 domain, central accessory domain and C-terminal catalytic domain. A previous study showed that the C2 domain of Vps34 is sufficient to bind UVRAG (Liang et al., 2006); thus, we determined which region of Vps34 Atg14 interacts with. Full-length FLAG-Vps34, FLAG-Vps34N (1-282 amino acids, containing the C2 domain), or FLAG-Vps34C (283-887) was coexpressed with HA-UVRAG and HA-Atg14 (Figure 2B). Immunoprecipitation analysis revealed that Atg14 as well as UVRAG interacts with the N-terminal C2 domain of Vps34, suggesting that one Vps34 molecule might interact with either Atg14 or UVRAG through the C2 domain. Beclin 1 was also specifically coprecipitated with Vps34N, as reported previously (Liang et al., 2006).
To analyze the endogenous Atg14 complex, we generated antibodies against human Atg14 and UVRAG. Anti-Atg14 antibody was able to precipitate endogenous Atg14, Vps34, and Beclin 1, but not UVRAG (Figure 2C). Likewise, endogenous Vps34 and Beclin 1, but not Atg14, were coprecipitated with anti-UVRAG antibody. These data confirm the interactions of endogenous proteins, which were shown in the transfection experiments.
We confirmed that the Atg14 protein complex also contained p150, which is a regulatory subunit of Vps34 (Figure 2D). All these data suggest that human Atg14 and UVRAG form two distinct PI3-kinase complexes with Beclin 1, Vps34, and p150.
Beclin 1 and Vps34 Are Important for Expression Levels of Atg14 and UVRAG
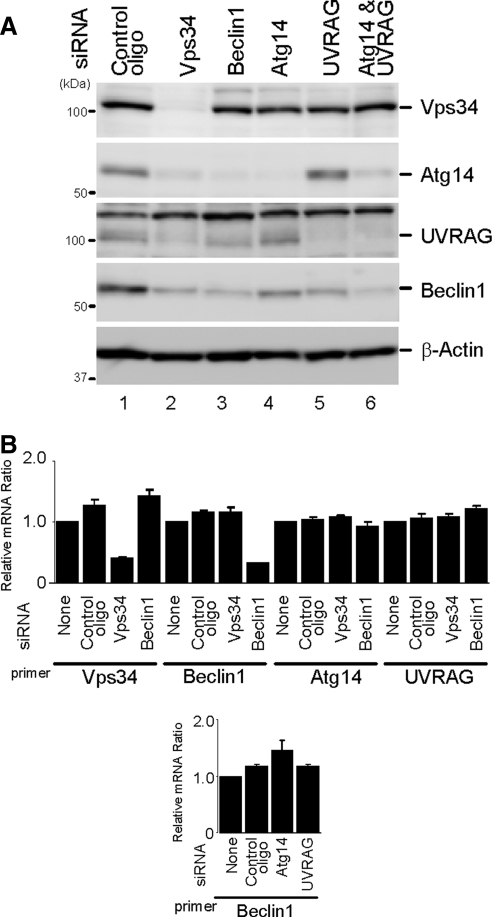
We next performed siRNA-mediated knockdown experiments using HeLa cells, because knockdown efficiency is very high in these cells. HeLa cells were transfected with specific siRNAs to knockdown endogenous Vps34, Beclin 1, Atg14, and UVRAG expressions. At 72 h after the transfection, respective siRNA, but not a control siRNA, significantly reduced endogenous expression of each protein (Figure 3A). In addition to these direct effects, treatment with Vps34 siRNA also decreased Beclin 1, Atg14, and UVRAG expression levels (Figure 3A, lane 2). Atg14 and UVRAG protein levels also decreased in cells treated with Beclin 1 siRNA (Figure 3A, lane 3). Furthermore, siRNA against either Atg14 or UVRAG also modestly decreased Beclin 1 protein levels (Figure 3A, lanes 4 and 5), and the knockdown of both Atg14 and UVRAG reduced the Beclin 1 expression level more markedly (Figure 3A, lane 6). Changes in the mRNA levels were only found in each specific siRNA target (Figure 3B). These results suggest that the presence of Beclin 1 and Vps34 is important for the stability of both Atg14 and UVRAG, and Beclin 1 stability depends on Vps34, Atg14, and UVRAG.
Figure 3.
Beclin 1 and Vps34 are important for expression levels of Atg14 and UVRAG. (A) HeLa cells were treated with the indicated siRNA. Total cell lysates were prepared and analyzed by immunoblotting using the indicated antibodies. (B) Effect of each siRNA on mRNA expression levels. HeLa cells were treated with the indicated siRNAs for 72 h, and mRNA level was measured by real-time PCR. Data are expressed as mean ± SE of six PCR reactions.
Atg14 Is Present on Isolation Membranes
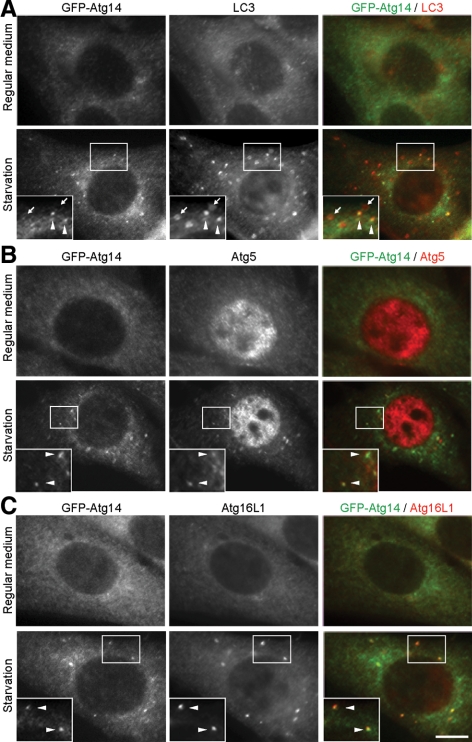
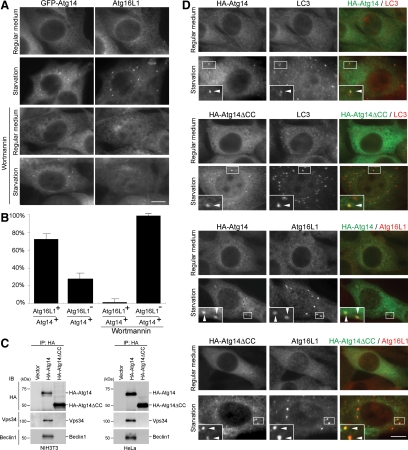
In yeast, Atg14 is present on PAS and is thought to function at the early stage of autophagosome formation. We therefore determined the intracellular localization of human Atg14. We used NIH3T3 cells stably expressing Atg14 N-terminally fused to GFP, because the numbers of isolation membrane and autophagosome are larger in NIH3T3 cells than in HEK293T and HeLa cells, which allowed us to perform morphological analysis more precisely. In these cells, although the GFP-Atg14 signal was primarily observed as a diffuse or reticular pattern in the cytoplasm, it was also detected as punctate dots, whose number increased during starvation (Figure 4A). These dots partially overlapped with LC3-positive dots, which represented autophagic vacuoles (Figure 4A, arrowheads, ∼25% of the total LC3 puncta). However, GFP-Atg14 was not present on most of the large LC3 dots (Figure 4A, arrows), raising the possibility that GFP-Atg14 might be present on only intermediate structures, but not on complete autophagosomes. Because the Atg12-Atg5 conjugate and Atg16L1 specifically localize on elongating isolation membranes (phagophore) (Mizushima et al., 2001), we tested their colocalization with Atg14. We observed better colocalization between GFP-Atg14 and endogenous Atg5, and between GFP-Atg14 and endogenous Atg16L1 after autophagy induction by starvation (Figure 4, B and C). Approximately 85% of Atg16L1 dots were positive for Atg14, and ∼75% of Atg14 dots were positive for Atg16L1. These data suggested that Atg14 primarily localizes on isolation membranes, but not on established autophagosomes.
Figure 4.
Atg14 localizes on isolation membranes. NIH3T3 cells stably expressing GFP-Atg14 were cultured in regular DMEM or amino acid-free DMEM without FBS for 2 h. Then, cells were fixed, permeabilized, and subjected to immunofluorescence microscopy by using rat anti-GFP antibody and either polyclonal anti-LC3 (A), anti-Atg5 (B), or anti-Atg16L1 (C) antibody. Structures positive for both Atg14 and LC3 are indicated by arrowheads, and structures positive for LC3 but negative for Atg14 are indicated with arrows. Bar, 10 μm.
PI3-Kinase Activity Is Not Required for Starvation-induced Atg14 Puncta Formation
To test whether Atg14 puncta formation depends on PI3-kinase activity, we used a PI3-kinase inhibitor wortmannin, which is known to inhibit autophagy (Blommaart et al., 1997). Wortmannin completely inhibited starvation-induced Atg16L1 puncta formation as described previously for the Atg5 puncta (Mizushima et al., 2001). However, starvation-induced Atg14 puncta remained in wortmannin-treated cells, suggesting that Vps34 activity is not required for Atg14 puncta formation (Figure 5, A and B).
Figure 5.
Atg14 puncta formation is independent of PI3-kinase activity. (A and B) Wortmannin inhibits puncta formation of Atg16L1 but not that of Atg14. NIH3T3 cells stably expressing GFP-Atg14 were cultured in regular DMEM or amino acid-free DMEM without FBS in the presence or absence of 200 nM wortmannin for 1 h. Then, cells were fixed, permeabilized, and subjected to immunofluorescence microscopy by using rat anti-GFP antibody and anti-rabbit Atg16L1 antibody. Bar, 10 μm (A). Atg16L1-positive rate of the Atg14 puncta under starvation conditions with or without wortmannin is shown as a percentage (B). Data are expressed as mean ± SD of at least 10 cells. (C) Atg14 interacts with Vps34 and Beclin 1 via the coiled-coil domain. Lysates were prepared from NIH3T3 cells stably expressing HA-Atg14 or HA-Atg14ΔCC and from HeLa cells transiently expressing HA-Atg14 or HA-Atg14ΔCC, and then they were subjected to immunoprecipitation analysis using anti-HA antibody. (D) Atg14 localizes on isolation membranes independently of binding to Vps34. NIH3T3 cells stably expressing HA-Atg14 or HA-Atg14ΔCC were cultured in regular DMEM or amino acid-free DMEM without FBS for 1 h. Then, cells were fixed, permeabilized, and subjected to immunofluorescence microscopy by using mouse anti-HA antibody and either polyclonal anti-LC3 or anti-Atg16L1 antibody. Structures positive for both markers are indicated by arrowheads. Bar, 10 μm.
We next examined localization of an Atg14 mutant unable to bind Vps34 and Beclin 1. Yeast Atg14 interacts with Vps34 and Vps30/Atg6 via the coiled-coil domain. We therefore designed a human Atg14 mutant (Atg14ΔCC), which lacks the coiled-coil domain (71-184 amino acid residues). In NIH3T3 and HeLa cells, both Vps34 and Beclin 1 were coprecipitated with Atg14, but not with Atg14ΔCC (Figure 5C), confirming that the coiled-coil region is important for the complex formation in mammals as well. Nonetheless, both wild-type Atg14 and Atg14ΔCC accumulated to punctate structures during starvation, which were colocalized with Atg16L1 and partially with LC3 (Figure 5D) These results suggest that Atg14 can localize to isolation membranes or its precursor structures independently of Vps34 and Beclin 1.
UVRAG Is Present on Rab9-positive Endocytic Compartments
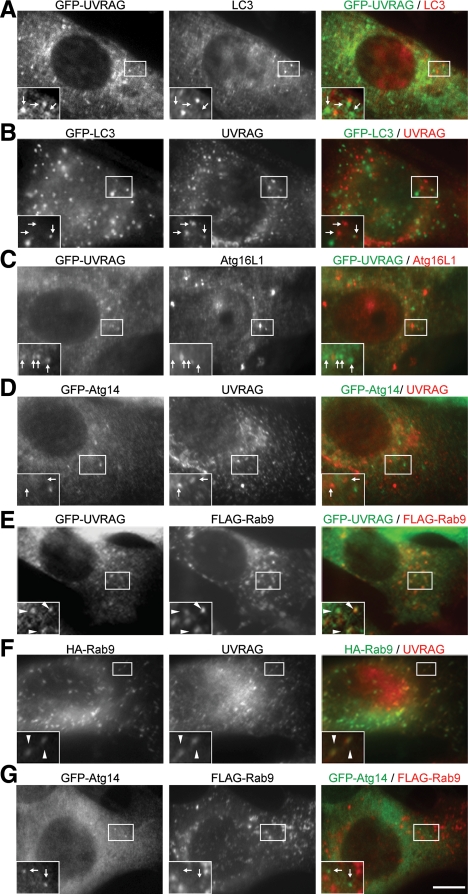
We next examined the intracellular localization of UVRAG. In NIH3T3 cells stably expressing UVRAG N-terminally fused to GFP (GFP-UVRAG), numerous dots were observed even under nutrient-rich conditions. Most of the GFP-UVRAG dots were distinct from LC3 (Figure 6A). Endogenous UVRAG was also not colocalized with GFP-LC3 even under starvation conditions (Figure 6B, starvation). We tested other autophagy factors such as Atg16L1 and Atg14, but failed to detect colocalization with UVRAG (Figure 6, C and D). These data suggest that these UVRAG dots do not represent autophagy-related structures. Considering the possibility that UVRAG might be a homolog of Vps38, we then analyzed the endocytic pathway. Among several marker proteins tested, UVRAG showed clear colocalization with Rab9 (∼80%), one of the late endosome markers (Figure 6, E and F). UVRAG also colocalized but only partially with other endosome markers such as Rab5 and Rab7 (Supplemental Figure 2). It was shown that Rab9 and Rab7 localize distinct type of late endosomes. Rab9-positive compartments contain mannose 6-phosphate receptors and the tail-interacting protein of 47-kDa (TIP47) and are involved in retrograde transport from late endosomes to TGN (Lombardi et al., 1993; Barbero et al., 2002). In contrast, Atg14 did not colocalize with Rab9 (Figure 6G). Therefore, UVRAG primarily resides in these endocytic compartments, not in the autophagy pathway.
Figure 6.
UVRAG localizes on Rab9-positive late endosomes. NIH3T3 cells (F) and NIH3T3 cells stably expressing either GFP-UVRAG (A, C, and E), GFP-LC3 (B), or GFP-Atg14 (D and G) were transiently transfected with FLAG-Rab9 (E and G), or HA-Rab9 (F) for 24 h. In the experiments in B, C, D, and G, cells were subjected to starvation by culturing in amino acid-free DMEM without FBS for 2 h before fixation. After fixation, the cells were subjected to immunocytochemistry by using rat anti-GFP (A–E and G), rabbit anti-LC3 (A), rabbit anti-Atg16L1 (C), rabbit anti-FLAG (E and G), mouse anti-HA (F), or rabbit anti-UVRAG (B, D and F) antibodies. Structures positive for both green and red signals are indicated by arrowheads, and those positive for either a green or red signal are indicated with arrows. Bar, 10 μm.
Vps34 Is Present on Rab9-positive Endocytic Compartments and Starvation-induced Atg14-positive Structure
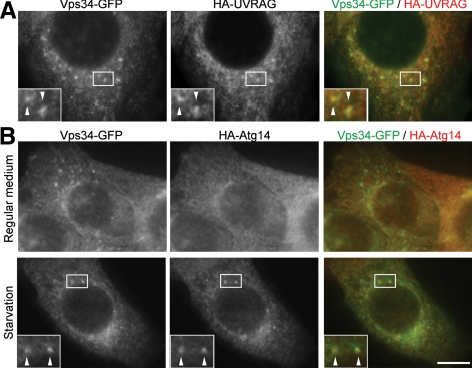
Because Vps34 interacts with both Atg14 and UVRAG, it is expected to localize to autophagic and endocytic Rab9-positive compartments. We generated Vps34 constructs tagged with GFP at its N terminus (GFP-Vps34) or C terminus (Vps34-GFP). Although GFP-Vps34 showed no punctate structures (data not shown), Vps34-GFP showed numerous dot structures under nonstarvation conditions (Figure 7A). These Vps34-GFP dots were colocalized with coexpressed HA-UVRAG almost completely. Under starvation conditions, ∼30% of Vps34-GFP dots were colocalized with coexpressed HA-Atg14 (Figure 7B). These results suggest that Atg14 and UVRAG function together with Vps34 in these compartments.
Figure 7.
Vps34 localizes on UVRAG-positive puncta and starvation-induced Atg14-positive puncta. NIH3T3 cells stably expressing Vps34-GFP and either HA-UVRAG (A) or HA-Atg14 (B) were generated. (B) Cells were subjected to starvation by culturing in amino acid-free DMEM without FBS for 2 h. After fixation, the cells were subjected to immunocytochemistry using rat anti-GFP and mouse anti-HA antibodies. Structures positive for both green and red signals are indicated by arrowheads. Bar, 10 μm.
Human Atg14 Is Required for Autophagosome Formation
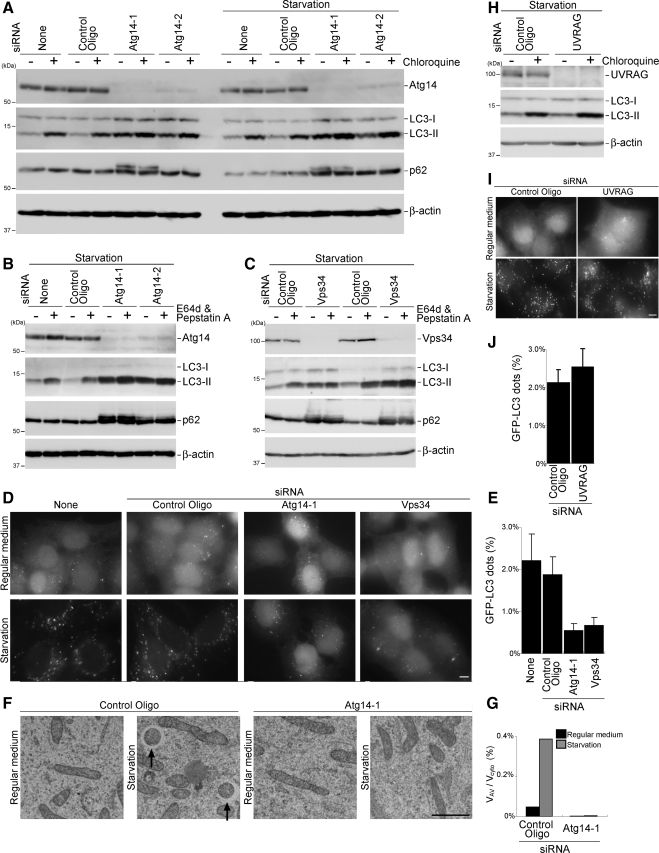
Although human Atg14 does not complement the yeast atg14 deletion mutant (Supplemental Figure 3A), subcellular localization analyses of Atg14 suggested that Atg14 functions in the autophagy pathway. To prove this hypothesis, we determined the effect of Atg14 silencing on autophagy in HeLa cells by using two sets of siRNA against different regions of Atg14 (Atg14-1 and Atg14-2). Atg14 protein levels decreased by nearly 100% and ∼90% after Atg14-1 and Atg14-2 siRNA treatments, respectively (Figure 8A). We then performed autophagy flux assay (Tanida et al., 2005; Mizushima and Yoshimori, 2007). In control cells, the amount of LC3-II increased after chloroquine treatment, which increased lysosome pH, under both nutrient-rich and starvation condition, suggesting the high autophagy flux in HeLa cells. In Atg14 siRNA-treated cells, LC3-II was still observed in rather higher levels. However, the LC3-II amount did not change after chloroquine treatment in Atg14-1 siRNA-treated cells and only slightly increased in Atg14-2 siRNA-treated cells (Figure 8A), suggesting that autophagic flux is blocked in these cells. We also monitored the expression level of autophagy substrate p62, another indicator of autophagic degradation (Bjørkøy et al., 2005; Mizushima and Yoshimori, 2007). The amount of p62 decreased after autophagy induction by starvation in control cells, which was inhibited in Atg14 siRNA-treated cells (Figure 8A). We obtained similar results when we used lysosome protease inhibitors E64d and pepstatin A, instead of chloroquine (Figure 8B). We also examined the effect of Vps34 silencing on autophagy. Vps34 siRNA caused accumulation of LC3-II under both nonstarvation and starvation conditions as did Atg14 siRNA treatment (Figure 8C). Again, E64d and pepstatin treatment did not lead to further accumulation of LC3-II in Vps34 siRNA-treated cells. Starvation-dependent p62 reduction was not also observed in these cells. All these data indicate that autophagy flux is suppressed in cells treated with siRNA against Atg14 and Vps34, although LC3 conversion is not inhibited.
Figure 8.
Atg14 and Vps34 are required for autophagosome formation. (A–C) Reduced autophagic flux in cells treated with Atg14 (A and B) and Vps34 siRNA (C). HeLa cells treated with the indicated siRNA were incubated in regular DMEM or starvation medium in the presence or absence of chloroquine (20 μM) for 1 h (A) or E64d (50 μM) and pepstatin A (50 μg/ml) for 2 h (B and C). Total lysates of the cells were analyzed by immunoblotting by using the indicated antibodies. (D and E) HeLa cells stably expressing GFP-LC3 were treated with Atg14 and Vps34 siRNA as described in A–C and starved for 2 h. Bar, 10 μm (D). The ratio of the total area of GFP-LC3 dots to the total cellular area in starved cells is shown as a percentage. Data are expressed as mean ± SD of at least 10 different fields (E). (F and G) HeLa cells treated with the control or Atg14 siRNA were starved for 2 h and subjected to conventional electron microscopic analysis. Autophagosomes are indicated by arrows. Bar, 1 μm (F). The ratio of total autophagosome area to total cytoplasmic area was determined by morphometric analysis. Ten cells were analyzed for each sample using MetaMorph image analysis software (G). (H) HeLa cells treated with UVRAG siRNA were incubated in starvation medium in the presence or absence of chloroquine (20 μM) for 1 h. (I and J) HeLa cells stably expressing GFP-LC3, which were treated with UVRAG siRNA, were starved for 2 h and GFP-LC3 puncta were observed. Bar, 10 μm (I). The ratio of the total area of GFP-LC3 dots to the total cellular area in starved cells is shown as a percentage. Data are expressed as mean ± SD of at least 10 different fields (J).
Finally, we determined at which step autophagy is perturbed. In HeLa cells stably expressing GFP-LC3, GFP-LC3 was mostly present in the nucleus under normal conditions, although its physiological significance is unknown (Kuma et al., 2007). GFP-LC3 was translocated to cytoplasmic dots when autophagy was normally induced (Figure 8D). However, the starvation induced GFP-LC3 puncta formation was markedly impaired in cells treated with siRNA against Atg14 and Vps34 (Figure 8, D and E). Instead, most GFP-LC3 remained in the nucleus in these cells. Electron microscopic analysis demonstrated that autophagosomes are virtually absent in Atg14 knockdown cells (Figure 8F). These fluorescence and electron microscopic analyses suggest that Atg14 is required for autophagosome formation rather than autophagosome maturation.
We also tested the effect of UVRAG knockdown on autophagy. In contrast to a previous report (Liang et al., 2006), we could not detect any inhibitory effect of UVRAG siRNA on autophagy flux and GFP-LC3 dot formation, although UVRAG expression was efficiently suppressed in the treated cells (Figure 8, H–J).
Coiled-Coil Domain of Atg14 Is Required for Autophagy
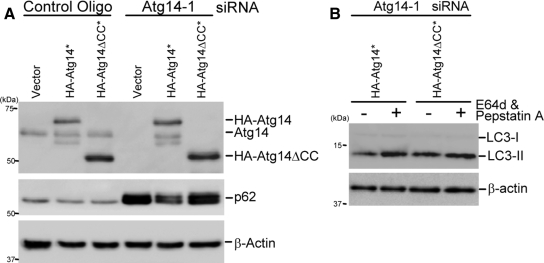
To investigate the role of Atg14 complex formation in autophagy, we analyzed the function of Atg14ΔCC, which is unable to interact with Beclin 1 and Vps34 (Figure 5C), by an Atg14 complementation assay. siRNA-resistant Atg14 and Atg14ΔCC were generated by introducing silent mutations in the siRNA target region, and they were expressed in Atg14-knockdown HeLa cells. In these cells, endogenous Atg14 was replaced with HA-Atg14 or HA-Atg14ΔCC (Figure 9A). Accumulation of p62 due to Atg14 depletion was restored by wild-type Atg14 but not by Atg14ΔCC (Figure 9A). LC3 flux was also restored by wild-type Atg14 but not by Atg14ΔCC (Figure 9B). Therefore, these data suggest that the coiled-coil domain of Atg14 is required for its involvement in autophagy probably through interaction with Vps34 and Beclin 1.
Figure 9.
Atg14ΔCC mutant cannot rescue the autophagy-defective phenotype of Atg14-knockdown cells. (A and B) HeLa cells were treated with the indicated siRNA. After 2 d, the cells were transiently transfected with an empty vector, HA-Atg14* (asterisk [*] indicates the siRNA-resistant version) or HA-Atg14ΔCC* plasmid together with the same siRNAs again, and then they were cultured for an additional 2 d. The cells were again transfected with the same expression vectors and cultured for another 1 d. Then, the cells were incubated in starvation medium for 2 h. Total lysates were prepared and p62 expression level (A) and LC3 turnover (B) were determined.
DISCUSSION
We concluded that KIAA0831, which was identified by a database search, is indeed a human Atg14 orthologue because it interacts with class III PI3-kinase complex subunits, is present on autophagic isolation membranes, and is required for autophagosome formation. The discovery of human Atg14 further provided lines of evidence showing that mammalian cells have at least two distinct class III PI3 kinase complexes as do yeast cells: the Atg14 complex (equivalent to yeast complex I) and the UVRAG complex (equivalent to yeast complex II). In contrast to human Atg14, UVRAG is mainly located on Rab9-positive endocytic compartments, suggesting that the function of these two distinct PI3-kinase complexes is different.
In yeast cells, autophagosomes are easily distinguished from PAS by fluorescence microscopy because autophagosomes can be enriched if autophagosome-vacuole fusion is inhibited, for example, by ypt7 mutation. However, because it is difficult to distinguish PAS from isolation membranes simply by fluorescence microscopy, it remains unknown whether yeast Atg14 is present on isolation membranes in addition to PAS. Our current results show the presence of human Atg14 on isolation membranes but not on established autophagosomes. The Atg12-Atg5 conjugate and Atg16L1 are currently used as isolation membrane markers (Mizushima et al., 2001). However, one part of the Atg14 punctate structures was negative for Atg16L1 (Figure 4C, see text). Furthermore, although Atg16L1 punctate formation was dependent on PI3-kinase activity, Atg14 punctate formation was not (Figure 5). These data suggest that Atg14 may localize to the isolation membrane or its precursor membrane (possibly equivalent to yeast PAS) earlier than Atg16L1.
We observed that LC3 lipidation occurs even in Atg14 or Vps34 siRNA-treated cells, even though autophagy is profoundly inhibited. Similar phenomenon was observed previously in Beclin 1-silenced mammalian cells (Zeng et al., 2006; Matsui et al., 2007) and FIP200 deficient cells (Hara et al., 2008). Yeast Atg8 lipidation also occurs in atg1, 2, 6, 9, 13, 14, 16, and 17 mutants (Suzuki et al., 2001), and vps15 and vps34 mutants (Obara et al., 2008a). Thus, LC3 conversion is not always an indicator of autophagy induction. As we performed in the present study, the autophagy flux assay would be more appropriate to measure the real autophagy activity. We do not know the reason why LC3-II rather accumulates in Atg14 and Vps34 siRNA-treated cells. LC3-II production on unknown aberrant membrane may impair LC3-II turnover.
Although autophagy is highly conserved from yeast to mammals, evolution of the Vps pathway is less clear. Functions of some Vps proteins such as ESCRT proteins seem to be highly conserved, but not all Vps proteins are present in higher eukaryotes. This is because the trafficking machinery between TGN and late endosomes is not identical in yeast and mammals. Although the involvement of Vps34 has been suggested in various membrane trafficking pathways such as the Golgi-to-lysosome trafficking pathway (Brown et al., 1995; Davidson, 1995), and internal vesicle formation in multivesicular late endosomes (Futter et al., 2001; Johnson et al., 2006), the role of Beclin 1 in these pathways has been controversial (Furuya et al., 2005; Takacs-Vellai et al., 2005; Zeng et al., 2006). In the present study, the role of complex II (the UVRAG complex) remains unknown. In a yeast complementation analysis, carboxypeptidase Y missorting in Δvps38 cells was restored by yeast Vps38 but not human UVRAG (Supplemental Figure 3B). Autophagy and liquid-phase endocytosis of dextran were not affected in cells treated with UVRAG siRNA as far as we examined (Figure 8 and data not shown, respectively). Complete suppression might be required to observe these functional effects. Nevertheless, the unique localization of UVRAG on Rab9-positive endosomes suggests that UVRAG exerts its functions in the endocytic pathways. UVRAG partially colocalizes with the early endosome marker Rab5, which may be consistent with a recent report that UVRAG functions together with the class C Vps complex (Liang et al., 2008). Although it has been reported that transiently overexpressed GFP-LC3 and UVRAG are colocalized (Liang et al., 2007), we did not observe such colocalization between endogenous LC3 and stably transfected GFP-UVRAG and between stably transfected GFP-LC3 and endogenous UVRAG (Figure 6, A and B).
Detailed studies of vps30 and vps38 mutant cells revealed that the primary defect in these mutants is failure to recruit the retromer complex to endosomes due to inability to produce endosomal PI(3)P. The retrograde defect inhibits recycling of Vps10, a receptor of the vacuolar protease carboxypeptidase Y (Burda et al., 2002). Considering that Rab9 is also involved in retrograde transport from late endosomes to TGNs (Lombardi et al., 1993; Barbero et al., 2002), the function of the type 2 class III PI3-kinase complex may be similar both in yeast and mammals.
Even if UVRAG is involved in retrograde transport or the endocytic pathway, it is still possible that UVRAG can regulate autophagy as reported previously (Liang et al., 2006). Retrograde transport may be important for normal recycling of other Atg proteins. It was reported that mammalian Atg9 cycles between TGN and endosomes, and part of Atg9 is present on the Rab9-positive compartment (Young et al., 2006). The redistribution of Atg9 from TGN to endosomes depends on ULK1, a mammalian Atg1 homologue, and PI3-kinase activity. The UVRAG complex may therefore regulate autophagy through mammalian Atg9 cycling. Another possibility is that these endosomes are required for autophagosome formation as membrane source, or for maturation into amphisomes (Berg et al., 1998).
The current study implies that there may be other types of class III PI3-kinase complex. For example, the class III PI3-kinase complexes including Beclin 1 were suggested to be involved in amino acid signaling upstream of mammalian target of rapamycin (Byfield et al., 2005; Nobukuni et al., 2005). Because this PI3-kinase complex positively regulates the amino acid signal, it seems totally opposite from its role in autophagy, which is up-regulated after amino acid withdrawal. Therefore, another specific subunit may be required for this specific function. Because there are several other processes that require PI3-kinase activity, detailed analysis of specific subunits would further facilitate our understanding of this evolutionally conserved system.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Tamotsu Yoshimori for providing the pCMV5 HA-Beclin 1, pcDNA3 FLAG-Vps34, and pcDNA3 MEF-p150 plasmids and the anti-LC3, anti-Beclin 1, and anti-Vps34 antibodies. We thank Dr. Mitsunori Fukuda for providing the pEF FLAG-Rab5A, pEF FLAG-Rab7A, and pEF FLAG-Rab9A plasmids; Dr. Toshio Kitamura for the retroviral vectors and Plat E cells; and Drs. Yoshinori Ohsumi and Keisuke Obara for the yeast Atg14 and Vps38 expression plasmids, Ape1 and CPY antibodies, and Δapg14 and Δvps38 strains. This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to N. M.) and grants for research fellowships from the Japan Society for the Promotion of Science for Young Scientists (to E. I.). We also thank the Kato Memorial Bioscience Foundation and Toray Science Foundation for financial support.
Abbreviations used:
- CVT
cytoplasm-to-vacuole targeting
- FBS
fatal bovine serum
- GFP
green fluorescent protein
- LC3
microtubule-associated protein light chain 3
- PAS
preautophagosomal structure
- PI3-kinase
phosphatidylinositol 3-kinase
- TGN
trans-Golgi network
- UVRAG
UV irradiation resistance-associated gene.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-01-0080) on October 8, 2008.
REFERENCES
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbero P., Bittova L., Pfeffer S. R. Visualization of Rab9-mediated vesicle transport from endosomes to the trans-Golgi in living cells. J. Cell Biol. 2002;156:511–518. doi: 10.1083/jcb.200109030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg T. O., Fengsrud M., Stromhaug P. E., Berg T., Seglen P. O. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J. Biol. Chem. 1998;273:21883–21892. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Øvervatn A., Stenmark H., Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommaart E. F., Krause U., Schellens J. P., Vreeling-Sindelarova H., Meijer A. J. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- Brown W. J., DeWald D. B., Emr S. D., Plutner H., Balch W. E. Role for phosphatidylinositol 3-kinase in the sorting and transport of newly synthesized lysosomal enzymes in mammalian cells. J. Cell Biol. 1995;130:781–796. doi: 10.1083/jcb.130.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P., Padilla S. M., Sarkar S., Emr S. D. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J. Cell Sci. 2002;115:3889–3900. doi: 10.1242/jcs.00090. [DOI] [PubMed] [Google Scholar]
- Byfield M. P., Murray J. T., Backer J. M. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J. Biol. Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- Cuervo A. M. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Davidson H. W. Wortmannin causes mistargeting of procathepsin D. Evidence for the involvement of a phosphatidylinositol 3-kinase in vesicular transport to lysosomes. J. Cell Biol. 1995;130:797–805. doi: 10.1083/jcb.130.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove S. K., et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y., Yoshimoto K., Ohsumi Y. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 2007;143:1132–1139. doi: 10.1104/pp.106.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya N., Yu J., Byfield M., Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy, and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- Futter C. E., Collinson L. M., Backer J. M., Hopkins C. R. Human VPS34 is required for internal vesicle formation within multivesicular endosomes. J. Cell Biol. 2001;155:1251–1264. doi: 10.1083/jcb.200108152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriely G., Kama R., Gerst J. E. Involvement of specific COPI subunits in protein sorting from the late endosome to the vacuole in yeast. Mol. Cell Biol. 2007;27:526–540. doi: 10.1128/MCB.00577-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J. L., Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E. H., Lewis M. J., Black M. W., Pelham H. R. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 2003;22:548–557. doi: 10.1093/emboj/cdg062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. E., Overmeyer J. H., Gunning W. T., Maltese W. A. Gene silencing reveals a specific function of hVps34 phosphatidylinositol 3-kinase in late versus early endosomes. J. Cell Sci. 2006;119:1219–1232. doi: 10.1242/jcs.02833. [DOI] [PubMed] [Google Scholar]
- Jones D. T., Swindells M. B. Getting the most from PSI-BLAST. Trends Biochem. Sci. 2002;27:161–164. doi: 10.1016/s0968-0004(01)02039-4. [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Kawamata T., Suzuki K., Ohsumi Y. Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2007;356:405–410. doi: 10.1016/j.bbrc.2007.02.150. [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka S., Okano T., Ohsumi M., Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- Kihara A., Kabeya Y., Ohsumi Y., Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Reports. 2001a;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Noda T., Ishihara N., Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 2001b;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang W.-P., Klionsky D. J. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and autophagy conjugation complex. J. Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- Komatsu M., et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Kuma A., Matsui M., Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–328. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Liang C., Feng P., Ku B., Dotan I., Canaani D., Oh B. H., Jung J. U. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat. Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Liang C., Feng P., Ku B., Oh B. H., Jung J. U. UVRAG: a new player in autophagy and tumor cell growth. Autophagy. 2007;3:69–71. doi: 10.4161/auto.3437. [DOI] [PubMed] [Google Scholar]
- Liang C., et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Liang X. H., Kleeman L. K., Jiang H. H., Gordon G., Goldman J. E., Berry G., Herman B., Levine B. Protection against fatal Sindbis virus encephalitis by Beclin, a novel Bcl-2-interacting protein. J. Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindmo K., Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J. Cell Sci. 2006;119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- Lombardi D., Soldati T., Riederer M. A., Goda Y., Zerial M., Pfeffer S. R. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 1993;12:677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Maiuri M. C., et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maria Fimia G., et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- Matsui Y., Takagi H., Qu X., Abdellatif M., Sakoda H., Asano T., Levine B., Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ. Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Kuma A., Kobayashi Y., Yamamoto A., Matsubae M., Takao T., Natsume T., Ohsumi Y., Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–667. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T. How to Interpret LC3 Immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- Nice D. C., Sato T. K., Stromhaug P. E., Emr S. D., Klionsky D. J. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 2002;277:30198–30207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobukuni T., et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. USA. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K., Noda T., Niimi K., Ohsumi Y., et al. Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae. Genes Cells. 2008a;13:537–547. doi: 10.1111/j.1365-2443.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- Obara K., Sekito T., Niimi K., Ohsumi Y. The ATG18-ATG2 complex is recruited to autophagic membranes via PtdIns(3)P and exerts an essential function. J. Biol. Chem. 2008b doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K., Sekito T., Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes–Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol. Biol. Cell. 2006;17:1527–1539. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., Packer M., Schneider M. D., Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Petiot A., Ogier-Denis E., Blommaart E. F., Meijer A. J., Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- Qu X., et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schu P. V., Takegawa K., Fry M. J., Stack J. H., Waterfield M. D., Emr S. D. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- Stack J. H., Herman P. K., Schu P. V., Emr S. D. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 1993;12:2195–2204. doi: 10.1002/j.1460-2075.1993.tb05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug P. E., Reggiori F., Guan J., Wang C. W., Klionsky D. J. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- Takacs-Vellai K., Vellai T., Puoti A., Passannante M., Wicky C., Streit A., Kovacs A. L., Muller F. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr. Biol. 2005;15:1513–1517. doi: 10.1016/j.cub.2005.07.035. [DOI] [PubMed] [Google Scholar]
- Tanida I., Minematsu-Ikeguchi N., Ueno T., Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Leevers S. J., Ahmadi K., Timms J., Katso R., Driscoll P. C., Woscholski R., Parker P. J., Waterfield M. D. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- Wei Y., Pattingre S., Sinha S., Bassik M., Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser A. E., Emr S. D. Novel PtdIns(3)P-binding protein Etf1 functions as an effector of the Vps34 PtdIns 3-kinase in autophagy. J. Cell Biol. 2002;158:761–772. doi: 10.1083/jcb.200112050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A. R., Chan E. Y., Hu X. W., Kochl R., Crawshaw S. G., High S., Hailey D. W., Lippincott-Schwartz J., Tooze S. A. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Yue Z., Jin S., Yang C., Levine A. J., Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Overmeyer J. H., Maltese W. A. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J. Cell Sci. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.