Abstract
Introduction
The pattern of exhaled breath volatile organic compounds represents a metabolic biosignature with the potential to identify and characterize lung cancer. Breath biosignature-based classification of homogeneous subgroups of lung cancer may be more accurate than a global breath signature. Combining breath biosignatures with clinical risk factors may improve the accuracy of the signature.
Objectives
Develop an exhaled breath biosignature of lung cancer using a colorimetric sensor array. Determine the accuracy of breath biosignatures of lung cancer characteristics with and without the inclusion of clinical risk factors.
Methods
The exhaled breath of 229 study subjects, 92 with lung cancer and 137 controls, was drawn across a colorimetric sensor array. Logistic prediction models were developed and statistically validated based on the color changes of the sensor. Age, sex, smoking history, and COPD were incorporated in the prediction models.
Results
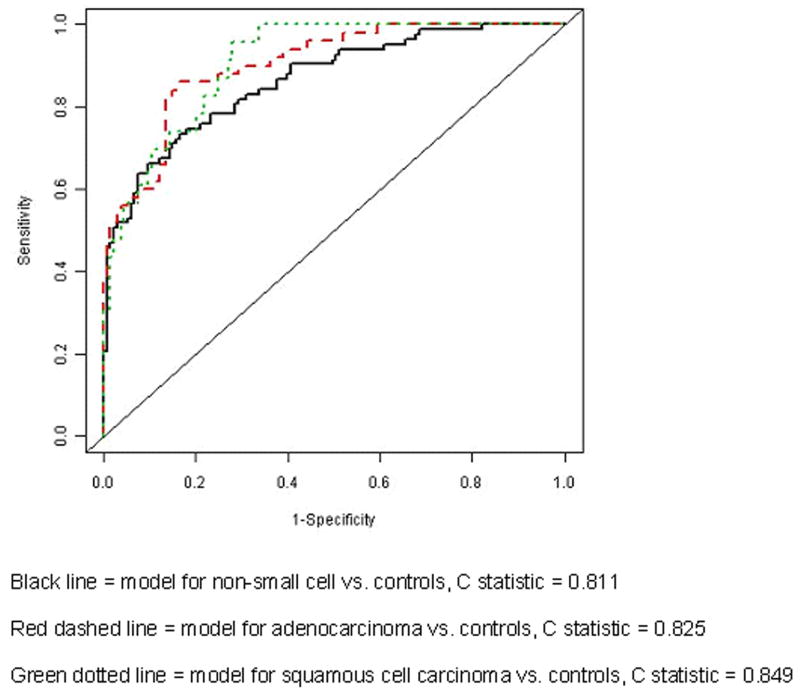
The validated prediction model of the combined breath and clinical biosignature was moderately accurate at distinguishing lung cancer from control subjects (C-statistic 0.811). The accuracy improved when the model focused on only one histology (C-statistic 0.825 – 0.890). Individuals with different histologies could be accurately distinguished from one another (C-statistic 0.889 for adenocarcinoma vs. squamous cell carcinoma). Moderate accuracies were noted for validated breath biosignatures of stage and survival (C-statistic 0.793, 0.770 respectively).
Conclusions
A colorimetric sensor array is capable of identifying exhaled breath biosignatures of lung cancer. The accuracy of breath biosignatures can be optimized by evaluating specific histologies and incorporating clinical risk factors.
Keywords: Breath analysis, biomarker, colorimetric sensor array
Introduction
The clinical evaluation and management of patients with lung cancer would benefit from the development of accurate, non-invasive, inexpensive biomarkers. Biomarkers capable of predicting the risk of developing lung cancer, identifying the presence of lung cancer, characterizing the nature of the cancer, and predicting and monitoring the response to therapy are being developed (1). These will lead to advances in primary prevention, chemoprevention, lung cancer screening, lung nodule management, lung cancer diagnosis, and the personalization of therapeutic choices.
Exhaled breath is an intriguing source of potential biomarkers of disease presence or activity. Volatile organic compounds (VOCs) are present in the exhaled breath in low concentrations. In principle, the composition of VOCs in the exhaled breath reflects metabolic activity within the body. Metabolic processes within the cells lead to the consumption and production of VOCs. These metabolic byproducts can circulate within the blood and transfer to the lungs where they are exhaled from the body. Thus, alterations in the body's metabolic processes may lead to unique breath VOC signatures.
There is evidence that lung cancer cells have unique metabolic properties (2-8). Evidence from the analysis of cell line headspace gas (9-12), as well as from the exhaled breath of lung cancer patients, suggests that this disease specific metabolism can be detected as breath signatures of the presence of lung cancer. The analysis of breath VOCs for lung cancer diagnosis has been performed with a variety of mass spectrometry techniques (13-22) as well as with various sensor arrays (23-29). Sensor arrays do not identify the specific constituents of exhaled breath; rather their output is the result of the interaction of the entire composition of the breath contents with the sensor. One such sensor device, called a colorimetric sensor array, is composed of chromogenic reagents printed on a disposable cartridge (30). The output from the sensor is a change in the colors of its elements. A previously reported study suggested that an early version of this sensor system was moderately accurate in identifying subjects with lung cancer based on their breath profile (23). Between that study and the study reported here, minor improvements were made to the colorimetric sensor platform, and the system was miniaturized (31).
Lung cancer is a heterogeneous disease, thus it is likely that there is more than one distinct lung cancer breath signature. Also, in other fields of lung cancer biomarker development, the accuracy of clinical and molecular risk predictors have been improved by combining the two approaches (32). The studies of breath analysis for lung cancer identification reported to date have not attempted to develop breath signatures related to the characteristics of the lung cancer (e.g. histology) or incorporated features of the study subjects into combined models. The aims of the current study were to: a) confirm the accuracy of a crude, portable colorimetric sensor array system for the detection of a lung cancer breath biosignature, b) determine the accuracy of breath biosignatures of lung cancer characteristics (histology, stage, survival), and c) determine the accuracy of breath biosignatures combined with relevant clinical variables.
Materials and Methods
Study design
This study was designed to determine if the responses of a colorimetric sensor array to the contents of exhaled breath are capable of identifying and characterizing lung cancer. Study subjects were recruited prospectively from outpatient clinics at the Cleveland Clinic. Their breath was sampled at a single point in time.
Patient population
The lung cancer study subjects all had biopsy proven, untreated lung cancer. The control group consisted of individuals at risk for developing lung cancer who were enrolled in a lung cancer screening study (age 40-75 years, 15+ pack-years of smoking, or 10+ pack-years of smoking with either COPD or a family history of lung cancer), and individuals with indeterminate lung nodules. The nature of the lung nodules was determined by biopsy or stability on CT surveillance with the length of follow-up determined by the clinician following the patient. Subjects with a prior lung cancer or other cancer within the past 5 years were excluded from enrollment, as were those requiring continuous supplemental oxygen, or receiving long-term immunosuppressive therapies. Recent food intake or cigarette smoking did not change a subject's study eligibility. The study protocol was approved by the Cleveland Clinic IRB and all study subjects signed informed consent.
Data collected on the lung cancer subjects included demographic information, co-morbid conditions (COPD was based on a clinical label or patient report), medications, and features of the cancer including the histology, stage, treatment, and survival. Information about survival was obtained through a review of the medical records, supplemented by the social security death index. Data collected on the at risk control group included demographic information, co-morbid conditions, and medications. Additional data collected on the lung nodule control group included the size of the nodule, how it was identified, and how it was evaluated.
Breath sampling and data processing
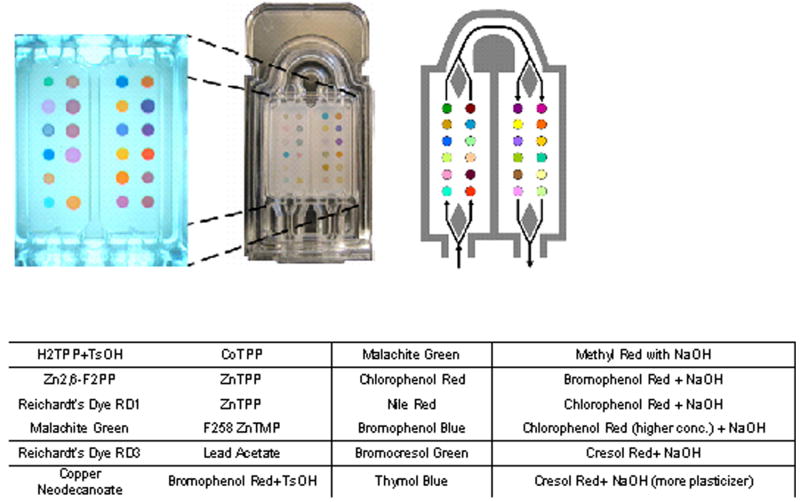
All study subjects performed tidal breathing, inhaling unfiltered air through their nose, and exhaling through their mouth into disposable corrugated tubing for a total of 5 minutes. Unpublished analysis of the colorimetric sensor responses suggested that equilibration of responses would be reached within 5 minutes. The exhaled breath was drawn across the sensor array at 200 ml/min by a pump placed distal to the array. The sensor array was composed of 24 separate colorants (Figure 1). Images of the array were taken at baseline and at 30 second intervals throughout breath collection. Each image of the sensor was converted to numerical values for changes in the red, green, and blue spectrum of each colorant. A new sensor array was used for each study subject. A separate array was exposed to room air (200 ml/min for 5 minutes) for each study subject, immediately prior to or after the subject's breath test was performed.
Figure 1.
Image of the colorimetric sensor used in the current study. 24 chemically reactive colorants are printed on a disposable cartridge. The reactive colorants used in the studied array are listed as they appear on the cartridge. Exhaled breath is drawn across the cartridge in the direction shown.
The colorimetric sensor array
The colorimetric sensor array approach (33) distinguishes among analytes or complex mixtures of analytes by its composite response; the array uses a diverse range of chemically responsive dyes, whose colors depend upon their chemical environment. The design of the disposable colorimetric sensor array used in this trial is based on dye-analyte interactions that are stronger than those that cause simple physical adsorption. The selected chemically-responsive dyes fall into three classes (described in detail previously (34,35)): (1) dyes containing metal ions (e.g., metalloporphyrins) that respond to Lewis basicity (that is, electron-pair donation, metal-ion ligation), (2) pH indicators that respond to Brønsted acidity/basicity (that is, proton acidity and hydrogen bonding), (3) dyes with large permanent dipoles (e.g., vapochromic or solvatochromic dyes) that respond to local polarity. This colorimetric sensor array, therefore, is responsive to the chemical reactivity of analytes, rather than to their effects on secondary physical properties (e.g., mass, conductivity, adsorption, etc.) as is generally the case with other electronic nose technology. The colorimetric sensor arrays generate high dimensional data (i.e., red, green and blue color changes for each dye; in these studies, 72 dimensional vectors), which allows for facile discrimination among even very complex mixtures (36). A picture of the array and list of the chemical dyes are shown in Figure 1. The sensitivity of the version of the colorimetric sensor array used in this study to chemical classes varied with the particular compound. Many relevant compounds were known to be detectable in the low parts per million range. The sensitivity to mixtures of compounds was less established.
Statistical analysis
The breath biosignatures of the following groups were compared:
Non-small cell carcinoma vs. all controls.
Adenocarcinoma vs. all controls.
Squamous cell carcinoma vs. all controls.
Adenocarcinoma vs. squamous cell carcinoma.
Small cell carcinoma vs. all controls.
Small cell carcinoma vs. non-small cell carcinoma.
Stage I and II non-small cell carcinoma vs. stage III and IV non-small cell carcinoma.
Survival < 12 months for all lung cancers vs. survival > 12 months for all lung cancers.
It is very difficult to estimate sample sizes for logistic models with multiple covariates as in this project, particularly when the number of discriminatory variables is unknown prior to the start of the project. To be comfortable that the confidence boundary for the estimates of sensitivity were within 10% of the stated value for the principal comparison (non-small cell carcinoma vs. all controls), the aim was to recruit a minimum of 50 subjects in the non-small cell cancer group and in the control group.
Four separate logistic prediction models were developed to distinguish between the groups of interest for each specific question. Totally 24 (the number of colorants) * 3 (changes in the red, green, and blue values) = 72 predictors were incorporated in the first logistic regression model for each specific question. A backwards step-down variable selection procedure was then performed. Under this approach, we started with fitting a model with all the variables of interest. The least significant variable was dropped, so long as it was not significant at our chosen critical level (the pre-specified significant level was p=0.05). We continued by successively re-fitting reduced models and applying the same rule until all remaining variables were statistically significant (37). We also checked distributional assumptions of the model to see if there were outliers, the observations that lie outside the overall pattern of the sample distribution. A similar approach was taken for the second model, wherein the 72 sensor predictors and 4 clinical predictors (age, sex, smoking status, and COPD) were included in the variable step-down procedure. In the third model, the 4 clinical predictors were forced into the model that included variables selected from the 72 breath predictors. The fourth model included only the 4 clinical variables. We validated the models statistically, using the bootstrapping method, and then calibrated the discrimination ability (38). The accuracy of each model is represented by the C-statistic for that model. The C-statistic is the area under an ROC curve, with 1.0 being an ideal test. Numerical data was compared using t-tests and categorical data was analyzed by Pearson's chi-square test procedure.
Results
Study subjects
229 subjects participated in this study, 92 with lung cancer and 137 controls. Of the 137 control subjects, 67 were subjects enrolled in a lung cancer screening trial, and 70 had indeterminate lung nodules (mean diameter 11 mm). Subjects with lung cancer had a higher mean age, lower portion of never smokers, and higher mean pack-years of cigarette use (Table 1). 83 of the 92 lung cancer subjects had non-small cell cancer, 50 of whom had an adenocarcinoma, and 23 a squamous cell carcinoma (Table 2). There was an equal distribution of stage I/II non-small cell cancers (41) and stage III/IV (42).
Table 1. Study subject details.
| All | Lung cancer | Control | P-Value | ||
|---|---|---|---|---|---|
| Subjects | 229 | 92 | 137 | ||
| Mean Age (years) | 62.9 | 68.9 | 58.9 | <0.001 | |
| Sex | F | 115 | 43 | 72 | 0.42 |
| M | 114 | 49 | 65 | ||
| Smoking | C | 53 | 25 | 28 | 0.006 |
| F | 129 | 58 | 71 | ||
| N | 44 | 9 | 35 | ||
| Mean # of Pyrs | 54 | 36 | <0.001 | ||
| COPD | 8 | 8 | 0.43 |
COPD = chronic obstructive pulmonary disease, F = female, M = male, C = current, F = former, N = never, Pyrs = pack-years
Table 2. Lung cancer features.
| Histology | Adenocarcinoma | 50 |
| Squamous cell | 23 | |
| Large cell | 3 | |
| Other non-small cell | 7 | |
| Small cell | 9 | |
| Stage | I | 32 |
| II | 9 | |
| III | 19 | |
| IV | 23 |
Accuracy of breath biosignature models
A series of breath biosignature models were developed for specific questions related to the identification and characterization of lung cancer. The groups compared for each question and the parameters considered when building the models are listed in the methods and in Table 3. The validated models, shown in Table 3, had lower accuracies then the initial models (mean decrease in C-statistic of 0.068). The models developed for the individual histologies compared to controls had higher accuracies than the model of the broader question of non-small cell cancer compared to controls (Tables 3, 4 and Figure 2). The accuracies of the models comparing the presence of cancer to controls were improved by including patient characteristics in a combined model (mean increase in C-statistic of 0.096). The accuracies of the models comparing features of the cancer (histology, stage, survival) were not influenced by including patient characteristics (mean increase in C-statistic of 0.002).
Table 3. Accuracy of statistically validated breath biosignature models.
| Groups Compared (n) | Model 1 | Model 2 | Model 3 | Model 4 | |
|---|---|---|---|---|---|
| Non-small cell (83) | Controls (137) | 0.701 | 0.811 | 0.761 | 0.710 |
| Adenocarcinoma (50) | Controls (137) | 0.784 | 0.747 | 0.825 | 0.695 |
| Squamous cell (23) | Controls (137) | 0.708 | 0.841 | 0.849 | 0.768 |
| Adenocarcinoma (50) | Squamous cell (22) | 0.889 | 0.742 | 0.864 | 0.517 |
| Small cell (9) | Controls (137) | 0.800 | 0.824 | 0.890 | 0.763 |
| Small cell (9) | Non-small cell (83) | 0.752 | 0.752 | 0.781 | 0.584 |
| Stages I and II (41) | Stages III and IV (42) | 0.792 | 0.793 | 0.784 | 0.460 |
| Survival < 12 months (24) | Survival > 12 months (68) | 0.768 | 0.761 | 0.770 | 0.576 |
The groups compared for each question are listed. This is followed by the C-statistic (area under the ROC curve) for the statistically validated models (model 1 = selected sensor parameters only, model 2 = selected sensor and selected clinical parameters, model 3 = selected sensor parameters and all 4 clinical parameters, model 4 = all 4 clinical parameters only).
Table 4. Representative sensitivity and specificity of the most accurate model for each study question.
| Groups Compared (n) | Model | Sensitivity | Specificity | |
|---|---|---|---|---|
| Non-small cell (83) | Controls (137) | 2 | 70 | 86 |
| Adenocarcinoma (50) | Controls (137) | 3 | 80 | 86 |
| Squamous cell (23) | Controls (137) | 3 | 91 | 73 |
| Adenocarcinoma (50) | Squamous cell (22) | 1 | 90 | 83 |
| Small cell (9) | Controls (137) | 3 | 89 | 85 |
| Small cell (9) | Non-small cell (83) | 3 | 78 | 95 |
| Stages I and II (41) | Stages III and IV (42) | 2 | 81 | 73 |
| Survival < 12 months (24) | Survival > 12 months (68) | 3 | 70 | 86 |
Figure 2.
ROC curves for the most accurate validated models comparing non-small cell carcinoma to controls, and the individual non-small cell carcinoma histologies to controls.
Discussion
The current study verified the potential of a colorimetric sensor array system to identify lung cancer breath biosignatures with a moderate accuracy for the distinction of subjects with lung cancer from a cohort of clinically relevant control subjects. This study suggests that the accuracy of breath biosignatures of specific lung cancer histologies is higher than that of a global cancer vs. no cancer signature. When distinguishing patients with and without lung cancer, a signature that combined breath and clinical predictors resulted in higher accuracy than either alone. The results also suggest that breath biosignatures can distinguish between lung cancers of different histology, and that characteristics of the lung cancer, such as stage and survival, can be predicted with breath analysis.
The improved accuracy of a biomarker based on metabolic changes, obtained by attempting to refine the application of the biomarker to a more homogeneous subgroup, is not surprising. Differences in the characteristics of individuals who develop specific lung cancer histologies, as well as differences in their clinical and imaging presentations, are well established. Recently, differences in the response to specific treatments between histologic groups (39), and the presence of unique molecular changes capable of further defining the nature of these groups (40), have been recognized. It is thus reasonable to speculate that metabolic differences exist between the cancer histologies that could lead to distinct VOC patterns in the breath. The implication of this finding for lung cancer breath test development is that a large number of well characterized lung cancer subjects' breath should be analyzed with biosignatures developed for specific patient subgroups. To maximize the accuracy of the broad question, looking for the presence of lung cancer in one individual, would thus require sequential decision making with analysis of a subjects' breath profile compared to a series of sub-group specific sensor profiles. The additional implication is that a breath biosignature may tell us more about the cancer than just whether or not it is present.
The ability to characterize a cancer's stage or prognosis with a metabolic biosignature is also not surprising. Rapid growth and widespread disease are likely to be characterized by distinct or magnified metabolic alterations. If these findings are supported by future, larger and more refined trials, this would imply that breath biosignatures may be capable of influencing decisions about the need for additional testing, the aggressiveness and type of treatment to offer, and be capable of monitoring the response to therapy.
As in other areas of biomarker development, the combination of clinical variables with molecular predictors was shown to improve overall test accuracy (32). Future breath test development should determine which clinical variables are independent of the breath signature and incorporate those that are complimentary to the overall accuracy of the test.
This study has several limitations related to its design and other limitations based on the nature of cross-reactive chemical sensors. The most obvious limitation relates to the breath collection technique. Breath was sampled in a controlled environment with coaching about the pace of breathing. All subjects underwent the same sampling procedure. However, attempts to isolate alveolar fractions of the breath, and to objectively measure or control the flow rate of exhaled breath were not used. These factors are known to influence the relative composition of breath analytes (41,42). In addition, the influence of diet was not studied (43), nor was the reproducibility of an individual subject's breath signature. Ambient air contains many volatiles which are present in the breath. Although breath testing was performed in a very limited number of locations without systematic selection of location based on disease, the influence of ambient volatiles cannot be ignored (44). To this end, models were developed using the difference between the sensor response to each subject's breath and the sensor response to ambient air. The models developed after correcting for ambient air were similar to those reported (data not shown). This method of correcting for the influence of exogenous volatiles, termed an alveolar gradient, is not likely to be appropriate for cross-responsive sensors where the influence on sensor response of a given element in the mixture is not linear. An alternative method to control for this influence is inhaling pure or filtered air for a period of time. Even this is not ideal as it is not possible to eliminate extremely low concentrations of potentially influential volatiles, and the time required for exogenous volatiles to leave the body is highly variable making the testing less practical. There were only minor improvements made to the colorimetric sensor array system used in this study compared to the first lung cancer study with this technology (31). The sensor used in this study was limited in its ability to detect all potentially relevant compounds at the very low concentrations found in the breath. Finally, a robust statistically validated model is reported, but validation on an independent cohort of subjects was not performed. Given these limitations, the results of this study should be interpreted as promising and able to provide direction for the design of future trials, but not as a definitive statement about the accuracy of breath testing with a colorimetric sensor array for lung cancer. It is reassuring that the accuracy of the breath biosignature for the broad question of lung cancer versus control reported here was similar to the prior study using this technology.
Another frequently described limitation of cross-reactive sensor systems is that they are not able to identify the individual components of a breath mixture. Instead, their response is representative of the entire mixture of breath chemicals. The strengths of these sensors are that they are capable of identifying a multi-dimensional discriminatory pattern of breath analytes, can be relatively inexpensive, can be used as a bedside test, and will not require advanced training for their use or interpretation. The weakness is that they cannot satisfy the need to explain the nature and origin of the breath analytes. Many complimentary technologies (gas chromatography-mass spectrometry, proton transfer reaction mass spectrometry, ion mobility spectrometry) are being used in an effort to identify the nature of these compounds (13-22). These technologies are more difficult to translate into inexpensive bedside tests. Acceptance of breath testing by the scientific and clinical communities will require the combination of advances in breath testing science provided by all methods of analysis.
Since this study was completed, a number of fundamental advances in the colorimetric sensor array technology have occurred (34,35). The most recent generation of colorimetric sensors uses robotic printing of reactive pigments in place of dyes, created by immobilizing chromogenic reagents in a nanoporous maxtrix of organically modified siloxanes. The resulting nanoporous matrix has far greater surface area for reactions to occur, dramatically improving the sensitivity of the sensor to all classes of relevant VOCs. These sensitivities have been further improved by imaging the sensor with enhanced optics and by extending the array from the 24 indicators used in the prior study to well over 100. Finally, a dedication to following principles of breath collection and delivery will allow us to determine the true potential of this biomarker moving forward. Additional work with complementary systems, capable of identifying the chemical composition of the unique breath constituents, will help us to discover the origin of the discriminatory VOCs, their movement into the breath, and the pathogenic processes that lead to the identified metabolic changes. This knowledge may help to improve our understanding of the nature of lung cancer and provide insights into novel methods of prevention and treatment.
In summary, the analysis of metabolic biomarkers in the exhaled breath of defined subgroups of lung cancer subjects with a colorimetric sensor array may allow us to identify and characterize lung cancer. The accuracy for identifying lung cancer can be optimized by combining clinical and breath predictors.
Acknowledgments
The authors thank Drs. Sung Lim and Brian Taba from iSense Medical Corp, in Redwood City, California, which is developing the next generation sensor, for assistance in image analysis.
Equipment for this project was provided by Chemsensing, Inc, Champaign, Ill.
Funding support was provided by the State of Ohio Third Frontier Program, ODOD TECH 06-55 and in part (KSS and JWK) through the NIH Genes, Environment and Health Initiative (U01ES016011)
References
- 1.Mazzone PJ. Give me a sign, any sign. Thorax. 2009;64:737–738. doi: 10.1136/thx.2008.107409. [DOI] [PubMed] [Google Scholar]
- 2.Goto I, Yoneda S, Yamamoto M, et al. Prognostic significance of germ line polymorphisms of the CYP1A1 and glutathione S-transferase genes in patients with non-small cell lung cancer. Cancer Res. 1996;56:3725–3730. [PubMed] [Google Scholar]
- 3.Ho JC, Zheng S, Comhair SAA, et al. Differential expression of manganese superoxide dismutase and catalase in lung cancer. Cancer Res. 2001;61:8578–8585. [PubMed] [Google Scholar]
- 4.Lauderoute KR, Amin K, Calaoagan JM, et al. 5-AMP-activated protein kinase (AMPK) is induced by low-oxygen and glucose deprivation conditions found in solid-tumor microenvironments. Molec Cell Biol. 2006;26:5336–5347. doi: 10.1128/MCB.00166-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel M, Lu L, Zander DS, et al. ALDH1A1 and ALDH3A1 expression in lung cancers: Correlation with histologic type and potential precursors. Lung Cancer. 2008;59:340–349. doi: 10.1016/j.lungcan.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 6.Jordan KW, Adkins CB, Su L, et al. Comparison of squamous cell carcinoma and adenocarcinoma of the lung by metabolomic analysis of tissue-serum pairs. Lung Cancer. 2010;68:44–50. doi: 10.1016/j.lungcan.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.An Z, Chen Y, Zhang R, et al. Integrated ionization approach for RRLC-MS/Ms-based metabonomics: Finding potential biomarker for lung cancer. J Proteome Res. 2010;9:4071–4081. doi: 10.1021/pr100265g. [DOI] [PubMed] [Google Scholar]
- 8.Duarte IF, Rocha CM, Barros AS, et al. Can nuclear magnetic resonance (NMR) spectroscopy reveal different metabolic signatures for lung tumours? Virchows Arch. 2010;457:715–725. doi: 10.1007/s00428-010-0993-6. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Xu F, Wang W, et al. A study of the volatile organic compounds exhaled by lung cancer cells in vitro for breath diagnosis. Cancer. 2007;110:835–844. doi: 10.1002/cncr.22844. [DOI] [PubMed] [Google Scholar]
- 10.Filipiak W, Sponring A, Mikoviny T, et al. Release of volatile organic compounds (VOCs) from the lung cancer cell line CALU-1 in vitro. Cancer Cell Int. 2008;8:17. doi: 10.1186/1475-2867-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sponring A, Filipiak W, Mikoviny T, et al. Release of volatile organic compounds from the lung cancer cell line NCI-H2087 in vitro. Anticancer Res. 2009;29:419–426. [PubMed] [Google Scholar]
- 12.Filipiak W, Sponring A, Filipiak A, et al. TD-GC-MS analysis of volatile metabolites of human lung cancer and normal cells in vitro. Cancer Epidemiol Biomarkers Prev. 2010;19:182–195. doi: 10.1158/1055-9965.EPI-09-0162. [DOI] [PubMed] [Google Scholar]
- 13.Phillips M, Altorki N, Austin JHM, et al. Prediction of lung cancer using volatile biomarkers in breath. Cancer Biomarkers. 2007;3:95–109. doi: 10.3233/cbm-2007-3204. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Xu F, Wang W, et al. A study of the volatile organic compounds exhaled by lung cancer cells in vitro for breath diagnosis. Cancer. 2007;110:835–844. doi: 10.1002/cncr.22844. [DOI] [PubMed] [Google Scholar]
- 15.Wehinger A, Schmid A, Mechtcheriakov S, et al. Lung cancer detection by proton transfer reaction mass-spectrometric analysis of human breath gas. J Mass Spectrom. 2007;265:49–59. [Google Scholar]
- 16.Peng G, Trock E, Haick H. Detecting simulated patterns of lung cancer biomarkers by random network of single-walled carbon nanotubes coated with nonpolymeric organic materials. Nano Letters. 2008;8:3631–3635. doi: 10.1021/nl801577u. [DOI] [PubMed] [Google Scholar]
- 17.Gaspar EM, Lucena AF, da Costa JD, et al. Organic metabolites in exhaled human breath – A multivariate approach for identification of biomarkers in lung disorders. J Chromatogr A. 2009;1216:2749–2756. doi: 10.1016/j.chroma.2008.10.125. [DOI] [PubMed] [Google Scholar]
- 18.Westhoff M, Litterst P, Freitag L, et al. Ion mobility spectrometry for the detection of volatile organic compounds in exhaled breath of lung cancer patients – Results of a pilot study. Thorax. 2009;64:744–748. doi: 10.1136/thx.2008.099465. [DOI] [PubMed] [Google Scholar]
- 19.Poli D, Goldoni M, Caglieri A, et al. Breath analysis in non small cell lung cancer patients after surgical tumour resection. Acta Biomed. 2008;79S1:64–72. [PubMed] [Google Scholar]
- 20.Song G, Qin T, Liu H, et al. Quantitative breath analysis of volatile organic compounds of lung cancer patients in China. Lung Cancer. 2010;67:227–31. doi: 10.1016/j.lungcan.2009.03.029. [DOI] [PubMed] [Google Scholar]
- 21.Ligor M, Ligor T, Bajtarevic A, et al. Determination of volatile organic compounds in exhaled breath of patients with lung cancer using solid phase microextraction and gas chromatography mass spectrometry. Clin Chem Lab Med. 2009;47:550–560. doi: 10.1515/CCLM.2009.133. [DOI] [PubMed] [Google Scholar]
- 22.Bajtarevic A, Ager C, Pienz M, et al. Noninvasive detection of lung cancer by analysis of exhaled breath. BMC Cancer. 2009;9:348. doi: 10.1186/1471-2407-9-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazzone PJ, Hammel J, Dweik RA, et al. Lung cancer diagnosis by the analysis of exhaled breath with a colorimetric sensor array. Thorax. 2007;62:565–568. doi: 10.1136/thx.2006.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Natale C, Macagnano A, Martinelli E, et al. Lung cancer identification by the analysis of breath by means of an array of non-selective gas sensors. Biosens Bioelectron. 2003;18:1209–1218. doi: 10.1016/s0956-5663(03)00086-1. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Cao M, Li Y, et al. A study of an electronic nose for detection of lung cancer based on a virtual SAW gas sensors array and imaging recognition method. Meas Sci Technol. 2005;16:1535–1546. [Google Scholar]
- 26.Machado RF, Laskowski D, Deffenderfer O, et al. Detection of lung cancer by sensor array analyses of exhaled breath. Am J Respir Crit Care Med. 2005;171:1286–1291. doi: 10.1164/rccm.200409-1184OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng G, Trock E, Haick H. Detecting simulated patterns of lung cancer biomarkers by random network of single-walled carbon nanotubes coated with nonpolymeric organic materials. Nano Letters. 2008;8:3631–3635. doi: 10.1021/nl801577u. [DOI] [PubMed] [Google Scholar]
- 28.Dragonieri S, Annema JT, Schot R, et al. An electronic nose in the discrimination of patients with non-small cell lung cancer and COPD. Lung Cancer. 2009;64:166–170. doi: 10.1016/j.lungcan.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 29.Peng G, Tisch U, Adams O, et al. Diagnosing lung cancer in exhaled breath using gold nanoparticles. Nature Nanotech. 2009;4:669–673. doi: 10.1038/nnano.2009.235. [DOI] [PubMed] [Google Scholar]
- 30.Janzen MC, Ponder JB, Bailey DP, et al. Colorimetric sensor arrays for volatile organic compounds. Anal Chem. 2006;78:3591–3600. doi: 10.1021/ac052111s. [DOI] [PubMed] [Google Scholar]
- 31.Kim P, Albarella JD, Carey JR, et al. Towards the development of a portable device for the monitoring of gaseous toxic industrial chemicals based on a chemical sensor array. Sens Actuators B. 2008;134:307–312. [Google Scholar]
- 32.Raji OY, Agbaje OF, Duffy SW, et al. Incorporation of a genetic factor into an epidemiologic model for prediction of individual risk of lung cancer: The Liverpool Lung Project. Cancer Prev Res. 2010;3:664–9. doi: 10.1158/1940-6207.CAPR-09-0141. [DOI] [PubMed] [Google Scholar]
- 33.Suslick KS. An Optoelectronic Nose: ‘Seeing’ Smells by Means of Colorimetric Sensor Arrays. MRS Bulletin. 2004;29:720–725. doi: 10.1557/mrs2004.209. [DOI] [PubMed] [Google Scholar]
- 34.Lim SH, Feng L, Kemling JW, et al. An Optoelectronic Nose for Detection of Toxic Gases. Nature Chem. 2009;1:562–567. doi: 10.1038/nchem.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng L, Musto CJ, Kemling JW, et al. A Colorimetric Sensor Array for Determination and Identification of Toxic Industrial Chemicals. Anal Chem. 2010;82:9433–9440. doi: 10.1021/ac1020886. [DOI] [PubMed] [Google Scholar]
- 36.Suslick BA, Feng L, Suslick KS. Discrimination of Complex Mixtures by a Colorimetric Sensor Array: Coffee Aromas. Anal Chem. 2010;82:2067–2073. doi: 10.1021/ac902823w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agresti A. Categorical Data Analysis. second. New York: Wiley-Interscience; 2002. [Google Scholar]
- 38.Harrell FE. Regression Modeling Strategies: With Applications to Linear Models, Logistic Regression, and Survival Analysis. New York: Springer; 2001. [Google Scholar]
- 39.Scagliotti GV, Parikh P, von Pawel J, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol. 2008;26:3543–51. doi: 10.1200/JCO.2007.15.0375. [DOI] [PubMed] [Google Scholar]
- 40.Eberhard DA, Giaccone G, Johnson BE. Biomarkers of response to epidermal growth factor receptor inhibitors in non-small-cell lung cancer working group: Standardization for use in the clinical trial setting. J Clin Oncol. 2008;26:983–994. doi: 10.1200/JCO.2007.12.9858. [DOI] [PubMed] [Google Scholar]
- 41.Miekisch W, Kischkel S, Sawacki A, et al. Impact of sampling procedures on the results of breath analysis. J Breath Res. 2008 doi: 10.1088/1752-7155/2/2/026007. [DOI] [PubMed] [Google Scholar]
- 42.Cope KA, Watson MT, Foster WM, et al. Effects of ventilation on the collection of exhaled breath in humans. J Appl Physiol. 2004;97:1371–1379. doi: 10.1152/japplphysiol.01034.2003. [DOI] [PubMed] [Google Scholar]
- 43.Pauling L, Robinson AB, Teranishi R, et al. Quantitative analysis of urine vapor and breath by gas-liquid partition chromatography. Proc Nat Acad Sci. 1971;68:2374–2376. doi: 10.1073/pnas.68.10.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dragonieri S, Schot R, Mertens BJA, et al. An electronic nose in the discrimination of patients with asthma and controls. J Allergy Clin Immunol. 2007;120:856–862. doi: 10.1016/j.jaci.2007.05.043. [DOI] [PubMed] [Google Scholar]