Abstract
Cystic fibrosis (CF) is the most common inherited lethal disease in Caucasians which results in multiorgan dysfunction. However, 85% of the deaths are due to pulmonary infections. Infection by Burkholderia cenocepacia (B. cepacia) is a particularly lethal threat to CF patients because it causes severe and persistent lung inflammation and is resistant to nearly all available antibiotics. In CFTR ΔF508 (ΔF508) mouse macrophages, B. cepacia persists in vacuoles that do not fuse with the lysosomes and mediates increased production of IL-1β. It is believed that intracellular bacterial survival contributes to the persistence of the bacterium. Here we show for the first time that in wild-type but not in ΔF508 macrophages, many B. cepacia reside in autophagosomes that fuse with lysosomes at later stages of infection. Accordingly, association and intracellular survival of B. cepacia are higher in CFTR-ΔF508 macrophages than in WT macrophages. An autophagosome is a compartment that engulfs nonfunctional organelles and parts of the cytoplasm then delivers them to the lysosome for degradation to produce nutrients during periods of starvation or stress. Furthermore, we show that B. cepacia downregulates autophagy genes in WT and ΔF508 macrophages. However, autophagy dysfunction is more pronounced in ΔF508 macrophages since they already have compromised autophagy activity. We demonstrate that the autophagystimulating agent, rapamycin markedly decreases B. cepacia infection in vitro by enhancing the clearance of B. cepacia via induced autophagy. In vivo, rapamycin decreases bacterial burden in the lungs of CF mice and drastically reduces signs of lung inflammation. Together, our studies reveal that if efficiently activated, autophagy can control B. cepacia infection and ameliorate the associated inflammation. Therefore, autophagy is a novel target for new drug development for CF patients to control B. cepacia infection and accompanying inflammation.
Keywords: autophagy, rapamycin, cystic fibrosis, host-pathogen interaction, Burkholderia cenocepacia, inflammation, macrophages
Introduction
Cystic fibrosis (CF) is the most common inherited lethal disease in Caucasians. It is caused by mutations in the cystic fibrosis transmembrane conductance regulator encoded by the CFTR gene encoding a membrane chloride transporter.1–3 The pathogenic factors in CF airway disease include defective innate antimicrobial activity, altered mucociliary clearance, abnormal submucosal gland function and overproduction of reactive oxygen species (ROS).4–6 Chronic inflammation is most central to CF pathogenesis as a consequence to pulmonary infections and leads to lung damage resulting in 85% of the deaths.7–10 Human and mouse CF airway epithelia are autophagy deficient and exhibit highly reduced autophagosome formation.11,12 Autophagy is a conserved physiological process that eliminates nonfunctional organelles and recycles cytosolic components for the generation of nutrients during periods of stress or starvation.13,14 Autophagy targets cytosolic long-lived proteins and organelles for lysosomal degradation in eukaryotic cells and plays a role in innate immunity.15 Autophagy has been linked to a variety of disease states, including cancer, myopathies, neurodegeneration, Crohn disease, infection and inflammation.13,16–18 Formation of autophagosomes depends on a lipid kinase signaling complex containing class III PI3K and two ubiquitin-like conjugation pathways that activates expansion of the pre-autophagosomal membrane.19,20 The Atg12-Atg5-Atg16L complex is attached to the nascent autophagosome and recruits Atg8—microtubuleassociated protein 1 light chain 3 (LC3)—which is expressed initially as an unprocessed form. Then, pro-LC3 is cleaved by Atg4 to generate an active form, LC3-I.21 LC3-I interacts with phosphatidylethanolamine (PE), yielding LC3-II.15,22,23 Therefore the transformation of LC3I to LC3II denotes autophagy stimulation and autophagosome formation. Subsequently, the Atg12-Atg5-Atg16L complex detaches from the formed autophagosome.23,24 This uncoating event enables the autophagosome to fuse with the lysosome. A small GTP binding protein Rab7 and the lysosomal associated membrane proteins 1 and 2 (LAMP1, LAMP2) are needed for this process.20,25–27 Many pharmacological agents have been reported to induce autophagy, such as rapamycin, an inhibitor of the mTOR pathway (mammalian target of rapamycin). The mTOR pathway is active in the presence of nutrients and negatively regulated by starvation or rapamycin and, under these conditions, autophagy is activated.28
Autophagy contributes to the control of a variety of bacterial and viral infections. For example, Group A Streptococcus that escapes from the endosome is targeted to the autophagosome, and Atg5 deletion delays its clearance.29,30
Similarly, during Listeria monocytogenes infection, bacterial listeriolysin-O toxin-mediated escape from phagosomes induces autophagy.30–32 Furthermore, a subset of Salmonella entericacontaining vacuoles is targeted to autophagosomes, and Atg5 deletion also allows for more bacterial survival.33 Interestingly, Mycobacteria-containing vesicles are targeted to autophagosomes, which decreases mycobacterial survival.34 The antimicrobial role of autophagy has been extended to viral infections such as human immunodeficiency virus (HIV).35,36
Burkholderia cenocepacia (B. cepacia) is an opportunistic, multidrug-resistant bacterium that infects CF patients leading to severe inflammation followed by destruction of the lung tissue, sometimes resulting in necrotizing pneumonia leading to patient death.37,38 Unfortunately, B. cepacia is resistant to essentially all antibiotics and thus impossible to treat. B. cepacia adopts an extracellular or intracellular lifestyle.39,40 The bacterium can survive within a variety of eukaryotic cells such as amoebae, epithelial cells and human macrophages.41–46
The B. cepacia-containing vacuole within macrophages delays acidification, does not assemble the NADPH-oxidase complex and fails to activate Rab7.43,47–49 These phenotypes are further exaggerated in CFTR-defective macrophages. However, little is known regarding the nature of the compartment harboring B. cepacia and the mechanism by which the bacteria delay its delivery to the lysosome for degradation.
During B. cepacia infection, abundant inflammatory cytokines such as IL-1β are detected in the bronchoalveolar lavage (BAL) of CF patients.50–58 IL-1β is primarily expressed as a precursor inactive molecule, which is later cleaved by caspase-1 to yield active 17-kDa IL-1β.59 The biological activities of IL-1β include promoting inflammatory responses and leukocyte infiltration.
Here we show that in WT macrophages a moderate number of B. cepacia-containing vacuoles are labeled with the specific autophagy marker LC3 within 2 h post-infection. B. cepacia containing vacuoles delay the fusion with the lysosome for several hours. Notably, B. cepacia decreases the expression of essential autophagy molecules. This B. cepacia-mediated effect is exacerbated in ΔF508 cells which are intrinsically defective in autophagy activity.11,12 In ΔF508 macrophages, B. cepacia-containing vacuoles do not fuse with the lysosomes and do not have autophagosome characteristics. We demonstrate that this defect is reversible since stimulation of autophagy with rapamycin decreases the bacterial burden in vitro and in vivo by accelerating the delivery of B. cepacia to the lysosome for degradation. Rapamycin treatment also dramatically decreases the recruitment of inflammatory cells to the lungs of infected CF mice. Taken together, our data provide a preponderance of evidence that B. cepacia exploits the already defective autophagy pathway in ΔF508 macrophages to establish infection. Stimulating autophagy activity with rapamycin restores the ability of ΔF508 macrophages to control B. cepacia infection and the associated inflammation. Therefore, our studies support the notion that pharmacological stimulation of autophagy will be beneficial for CF patients to prevent B. cepacia infection and thwart the detrimental inflammatory response within the lungs of CF patients.
Results
Macrophages harboring the CFTR ΔF508 mutation support increased B. cepacia survival and produce more IL-1β than WT macrophages.
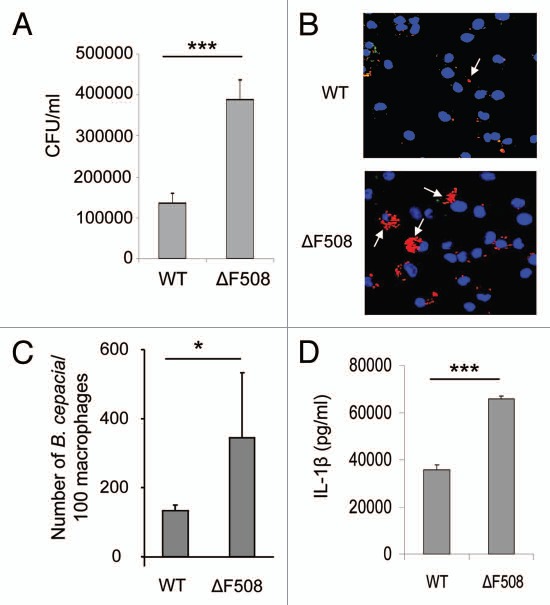
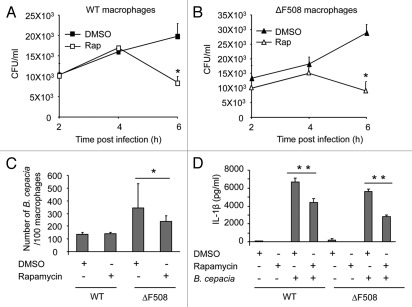
We examined whether B. cepacia had a survival advantage in primary murine macrophages expressing the CFTR protein harboring the ΔF508 mutation, which is the most common mutation in CF patients.60–62 WT and CFTR ΔF508 (ΔF508) macrophages were infected with the B. cepacia clinical isolate K56-2 and colony-forming units (CFU) were determined from lysed infected macrophages at 30 min (Fig. S1) and at 24 h post-infection (Fig. 1A). We found that more B. cepacia was recovered from ΔF508 macrophages than WT cells after 24 h of infection (Fig. 1A), whereas, the initial uptake was similar in both cells (Fig. S1). We next examined the number of B. cepacia associated with WT and ΔF508 macrophages by confocal microscopy. WT and ΔF508 macrophages were infected with red fluorescent protein (mRFP)-expressing B. cepacia for 30 min and 2 h and the number of B. cepacia associated with 100 macrophages was evaluated. At an early time point (30 min post-infection) similar numbers of B. cepacia were associated with WT and ΔF508 macrophages (Fig. S2). In contrast, at 2 h there were <200 B. cepacia associated with 100 WT macrophages (Fig. 1B and C), whereas there were >300 B. cepacia associated with 100 ΔF508 macrophages (Fig. 1B and C). Thus, these data are consistent with the CFU data suggesting more growth of B. cepacia in ΔF508 macrophages than in WT macrophages.
Figure 1.
Macrophages harboring ΔF508 mutation show more B. cepacia recovery and enhanced cytokine production from macrophages. (A) Wild-type (Wt) or CFTRΔF508 (ΔF508) murine bone marrow-derived macrophages (BMDM) were infected with B. cepacia and colony-forming units (CFUs) were enumerated at 24 h post-infection. (B) confocal microscopy of WT or ΔF508 BMDM after 2 h of infection with B. cepacia. Nuclei are stained blue with DAPI and B. cepacia expresses red florescent protein. White arrows indicate the sites of B. cepacia. (C) scoring of the number of B. cepacia associated with 100 BMDMs in confocal images. (D) 24 h post-infection, supernatants from BMDM from WT or ΔF508 mice were analyzed for IL-1β. Data are representative of 3 independent experiments and presented as the means ± SD. Asterisks in (C and E) indicate significant differences (*p < 0.05 and ***p < 0.0001).
Since IL-1β is an essential pro-inflammatory cytokine that affects CF patients,50–58 we next determined the levels of active IL-1β in culture supernatants and found that ΔF508 macrophages produced significantly more IL-1β when infected with B. cepacia compared with WT cells (Fig. 1D). Yet, the mechanism is unclear. To rule out the role of macrophage survival in IL-1β production differences, the release of lactate dehydrogenase (LDH) was evaluated in WT and ΔF508 macrophages infected with B. cepacia. The ΔF508 mutation did not alter macrophage survival in response to B. cepacia (Fig. S3). Therefore, primary macrophages expressing the ΔF508 mutation support increased B. cepacia intracellular survival and bacterial replication and produce more IL-1β during B. cepacia infection.
The B. cepacia-containing intracellular vacuole acquires autophagy characteristics in WT macrophages but not in ΔF508 macrophages.
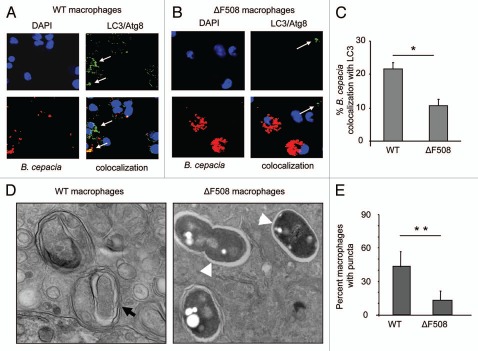
Since autophagy activity is compromised in CF epithelial cells we examined whether macrophages have the same defect by observing their autophagy response during the B. cepacia infection.11,12 WT and ΔF508 macrophages were infected with mRFP-expressing B. cepacia for 2 h. The acquisition of endogenous LC3 by the B. cepacia-containing vacuole was assessed with specific antibodies by confocal microscopy. In WT macrophages, 20% B. cepacia-containing vacuoles were labeled with the specific autophagy marker LC3 within 2 h infection (Fig. 2A and C). Several LC3-labeled vacuoles (puncta) were identified in WT macrophages (Fig. 2A; white arrows). In contrast, B. cepacia-containing vacuoles in ΔF508 macrophages had rare LC3-labeled structures (puncta) in response to B. cepacia compared with WT macrophages and only 10% of mRFP-expressing B. cepacia-containing vacuoles showed co-localization with LC3 (Fig. 2B and C). Notably, as shown earlier, greater numbers of B. cepacia were visualized in ΔF508 macrophages than in WT cells at 2 h post-infection (Fig. 2A and B). To further confirm the identity of the B. cepacia-containing compartment, B. cepacia-infected WT and ΔF508 macrophages were examined by transmission electron microscopy. Infected WT macrophages contained few B. cepacia and they were surrounded by several multilamellar membranes (Fig. 2D; black arrows) similar to autophagosomes and showed signs of bacterial degradation. In contrast, in ΔF508 macrophages, several intact B. cepacia were associated with the macrophage (Fig. 2D; white arrow heads), and they lacked the autophagosome-like structure. Immunofluorescence quantification of puncta within infected macrophages also confirmed that the autophagy response is compromised in ΔF508 macrophages during B. cepacia infection (Fig. 2E). Together, these data indicate that the autophagy response of ΔF508 macrophages to B. cepacia is significantly less than that of WT macrophages and hence more B. cepacia are enclosed in autophagosome-like vacuoles within WT macrophages.
Figure 2.
B. cepacia resides in an autophagosome-like compartment in macrophages. (A and B) confocal microscopy for wild-type (WT) murine BMDM (A) and BMDM harboring ΔF508 mutation (ΔF508) (B) representing the colocalization of B. cepacia with LC3/Atg8I after 2 h infection. Nuclei are stained with DAPI, B. cepacia expresses red florescent protein and LC3/Atg8 is stained green with specific antibodies. White arrows indicate the presence of LC3/Atg8. (C) the percentage of colocalization of B. cepacia with LC3/Atg8 was scored. (D) transmission electron microscopy of murine BMDM expressing WT or mutated ΔF508 CFtr protein after 2 h infection with B. cepacia. Black arrow indicates a mutilamellar autophagosome-like vacuole. White arrow heads indicate vacuoles containing intact bacteria within single-membrane vacuoles. (E) the scores for the percentage of puncta in 100 WT and ΔF508 macrophages after 4 h infection with B. cepacia. Data are representative of 3 independent experiments and presented as the means ± SD. Asterisks in (C and E) indicate significant differences (*p < 0.05; **p < 0.01).
B. cepacia downregulates autophagy genes during infection of WT and ΔF508 macrophages.
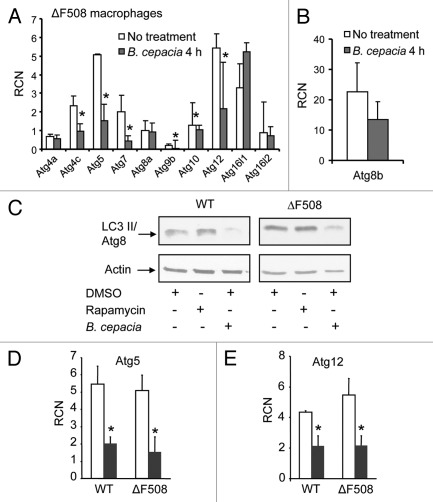
B. cepacia resides within LC3 labeled compartments reminiscent of autophagosomes in WT macrophages with the presence of puncta in several macrophages (Fig. 2). However, compared with WT macrophages infected with other organisms such as Salmonella, there were many fewer puncta in WT macrophages infected with B. cepacia (Fig. 2E and data not shown).13,14,63 Nevertheless, significantly fewer puncta were detected in ΔF508 macrophages when compared with WT macrophages infected with B. cepacia (Fig. 2E). The cause of this observation is unknown. To examine the effect of B. cepacia on the autophagy pathway, we performed an array analysis for a part of autophagy genes in B. cepacia infected WT and ΔF508 macrophages. B. cepacia infection led to a significant downregulation of several autophagy genes such as Atg9b, Atg5, Atg12 (Fig. 3A) and Atg8 (Fig. 3B) in ΔF508 and in WT macrophages (Fig. S4A and B). Quantitative PCR (qPCR) and protein gel blot analysis confirmed the downregulation of autophagy genes and their corresponding protein in both cell types (Figs. 3A, C–E and 2E and data not shown). The low number of puncta in ΔF508 macrophages compared with WT macrophages further confirmed that B. cepacia downregulates autophagy and that this effect is more profound in ΔF508 macrophages (Fig. 2E). On the other hand, other autophagyinteracting bacteria such as Salmonella induce the expression of several autophagy molecules during infection (data not shown). Therefore, it appears that the downregulation of essential autophagy molecules is a strategy employed by B. cepacia to delay the maturation of autophagosomes and hence avoid clearance by the host cell. The B. cepacia-mediated downregulation of autophagy genes is similar in WT and ΔF508 macrophages but the effect is more pronounced in the latter cells suggesting that they harbor an inefficient autophagy system as described in ΔF508 epithelial cells.11,12
Figure 3.
B. cepacia downregulates the expression of autophagy genes in murine macrophages. (A and B) Bone marrow-derived macrophages (BMDMs) harboring the ΔF508 mutation were either uninfected (No treatment) or infected with B. cepacia for 4 h and then the cells were lysed in trizol. RNA was extracted and SA Biosciences autophagy array analysis was performed for several autophagy genes (A) and for LC3/Atg8 (B). (C) Immunoblotting for WT and ΔF508 murine macrophages showing the expression level of LC3/Atg8 before and after 4 h infection with B. cepacia in the presence of rapamycin or DMSO. (D and E) q-PCR results for autophagy regulating genes 5 and 12 (Atg5, Atg12) in WT and ΔF508 BMDMs non infected (white bars) or infected (gray bars) with B. cepacia for 4 h. Data in (A, D and E) are expressed as relative copy numbers (RCN) and presented as means of 3 independent experiments ± SD. Asterisks indicate significant differences from the uninfected cells at the indicated time point (*p < 0.05).
Depletion of LC3 from WT macrophages allows for more B. cepacia recovery and increased IL-1β production.
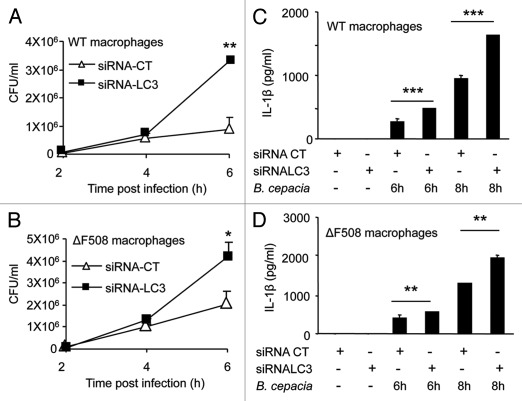
B. cepacia infection downregulates autophagy genes in both WT and ΔF508 macrophages, but the effect on autophagy activity is more pronounced in ΔF508 macrophages, suggesting a possible mechanism allowing for increased B. cepacia survival. This could be due to defective autophagy activity in macrophages harboring mutant CFTR protein similar to epithelial cells expressing the same mutation.11,12 Therefore, we examined if compromising autophagy in WT macrophages would result in a similar phenotype to ΔF508 macrophages. We reduced autophagy activity in WT macrophages by depleting LC3, an essential autophagy molecule. The depletion of LC3 was achieved with specific small interfering RNA (siRNA) to LC3 and confirmed by protein gel blots using specific antibody that recognizes LC3I and LC3II forms (Fig. S5A). Initial uptake of B. cepacia was similar in WT and ΔF508 macrophages pretreated with siRNA-LC3 or siRNA-control (CT) (data not shown). Depletion of LC3 in WT macrophages by specific siRNA was accompanied by an increased recovery of B. cepacia (Fig. 4A). Depletion of LC3 in ΔF508 macrophages was accompanied by an increase in B. cepacia recovery although less significant (Fig. 4B). The decrease in LC3 expression was associated with increased secretion of the active form of IL-1β from B. cepacia-infected WT and ΔF508 macrophages (Fig. 4C and D). Therefore, compromising autophagy in WT macrophages with specific siRNA produced a ΔF508-like phenotype, such as increased B. cepacia recovery and enhanced IL-1β release.
Figure 4.
Depletion of LC3 with specific siRNA increases B. cepacia multiplication and Interleukin-1β (IL-1β) secretion in macrophages. (A and B) Bone-marrow derived macrophages (BMDMs) from WT and ΔF508 mutant mice were nucleofected with siRNA against LC3/Atg8 (siRNA-LC3) or control siRNA (siRNA-CT) for 48 h and then infected with B. cepacia for 2, 4 and 6 h. colony forming units (CFUs) were enumerated. (C and D) Both types of macrophages were then infected for 6 and 8 h and supernatants were analyzed for IL-1β production. Data are representative of three different experiments and presented as the means ± SD. Asterisks indicate significant differences from the (siRNA-CT) at the respective time points (*p < 0.05; **p < 0.01; ***p< 0.001).
Once activated, caspase-1 cleaves pro-IL-1β to its active form, which is then released into culture supernatants. Therefore, to understand why IL-1β release is increased in ΔF508 macrophages during B. cepacia infection, we compared the levels of IL-1β pro-form in ΔF508 and WT macrophages upon B. cepacia infection. The depletion of LC3 with specific siRNA in WT and ΔF508 macrophages increased the levels of pro-IL-1β in macrophages in response to B. cepacia infection (Fig. S5B) and led to more active IL-1β released into culture supernatants (Fig. 4C and D). Therefore, decreasing autophagy activity resulted in increased pro-IL-1β accumulation, activation and release of active IL-1β in response to B. cepacia infection.
Stimulation of autophagy by rapamycin decreases B. cepacia recovery and IL-1β production in vitro.
Macrophages harboring ΔF508 mutation show increased intracellular survival and growth of B. cepacia (Fig. 1A and B). To investigate whether this phenotype is due to reduced autophagy activity in ΔF508 macrophages, we examined the effect of autophagy stimulation by rapamycin on B. cepacia-infected ΔF508 macrophages. Rapamycin is an inhibitor of the mammalian target of rapamycin (mTor) that induces autophagy.64,65 Macrophages were treated with rapamycin or with the drug vehicle DMSO before and after B. cepacia infection. The number of B. cepacia recovered was evaluated at 2, 4 and 6 h post infection and expressed as CFUs/ml. Rapamycin significantly decreased the number B. cepacia recovered from WT and ΔF508 macrophages (Fig. 5A and B). However, the effect was observed in ΔF508 macrophages as early as 2 h post-infection despite equal initial uptake (Fig. 5B and data not shown). Adding rapamycin directly to bacterial cultures in the absence of macrophages did not alter B. cepacia survival (data not shown). By confocal microscopy, the number of B. cepacia associated with 100 macrophages at 2 h post infection was significantly lower in ΔF508 macrophages in the presence of rapamycin, thus confirming the CFU data (Fig. 5C). In contrast, the reduction in B. cepacia numbers recovered from infected WT macrophages was evident at later stages of infection (Fig. 5A).
Figure 5.
Rapamycin treatment decreases the recovery of B. cepacia and Interleukin-1β (IL-1β) production from infected macrophages. (A and B) Bone-marrow derived macrophages (BMDMs) from WT mice (A) and those harboring the ΔF508 mutation (B) were infected with B. cepacia for 2, 4 and 6 h in the presence of rapamycin or DMSO. Colony-forming units (CFUs) were enumerated. (C) WT and ΔF508 were infected with B. cepacia for 2 h in the presence of rapamycin or DMSO and then the number of bacteria per 100 macrophages was scored. (D) WT and ΔF508 were infected with B. cepacia for 24 h in the presence of rapamycin or DMSO and the supernatants were analyzed for IL-1β. Data are representative of 3 independent experiments and presented as means ± SD. Asterisks indicate significant differences from the DMSO-treated cells (*p < 0.05; **p < 0.01).
We next investigated if rapamycin affects the levels of active IL-1β released in culture supernatants.66 In contrast to DMSO, rapamycin treatment of ΔF508 macrophages markedly decreased IL-1β production in response to B. cepacia infection (Figs. 5D and S6A). Rapamycin also decreased IL-6 production (Fig. S6B and C). The levels of other cytokines such as TNFα were comparable in the presence or absence of rapamycin at 24 h post-infection (Fig. S6D and E). Therefore, stimulation of autophagy by rapamycin decreases the production of IL-1β and not all pro-inflammatory cytokines in response to B. cepacia infection in vitro.
Rapamycin treatment decreases the bacterial burden and reduces the signs of inflammation in the lungs of ΔF508 mice.
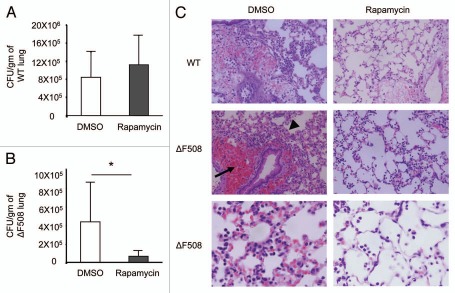
Since B. cepacia infects the lungs of CF patients we used the ΔF508 mouse model for CF to examine if rapamycin treatment decreases the bacterial burden in vivo and if it alleviates inflammatory findings in the lungs of infected mice. WT and ΔF508 mice were pretreated with two doses of rapamycin or DMSO by intra-peritoneal injections. Mice were then infected intra-tracheally with B. cepacia followed by an additional dose of rapamycin or DMSO. On the second day post-infection, mice were sacrificed, lungs were collected and the bacterial burden was determined. Rapamycin treatment did not affect B. cepacia recovery from WT lungs but reduced it drastically from ΔF508 lungs (Fig. 6A and B). Therefore, rapamycin treatment stimulates autophagy and decreases the B. cepacia burden in vivo in ΔF508 mice.
Figure 6.
Treatment with rapamycin markedly decreases the recovery of B. cepacia from infected lungs of mice harboring ΔF508 mutation in vivo. Wild-type (WT) (A) and mice harboring the ΔF508 mutation (ΔF508) (B) were pretreated with 2 doses of rapamycin (4 mg/kg) or with DMSO at a 24 h interval by intra-peritoneal injections. then, mice were infected intra-tracheally with B. cepacia followed by a dose of rapamycin or DMSO. colony-forming units (CFUs) recovered from homogenized lungs were enumerated and expressed as CFU per gram of lung tissue (A and B). (C) h&e staining of lung sections from WT (upper parts X40) or ΔF508 mice (middle parts X40) treated as in (A and B). Lower part shows higher magnification (X100) of infected ΔF508 lung sections. Data in (A and B) are represented as the means of data obtained from 3 mice ± SD. Asterisks indicate significant differences from the DMSO treated mice (*p < 0.05).
Next, we determined the effect of rapamycin treatment on the inflammatory response of lung tissue of B. cepacia-infected mice. Histological examination of H&E stained lung sections of WT mice infected with B. cepacia revealed the preservation of most of the lung tissue with patchy areas of accumulation of inflammatory cells (Fig. 6C and left upper part). Lung sections of ΔF508 mice demonstrated significantly more pronounced and generalized recruitment of inflammatory cells (arrow head) throughout the lung tissue with peribronchiolar and perivascular infiltrates and hemorrhage (arrow) (Fig. 6C and left middle part). Higher magnification of ΔF508 lung sections closely showed that B. cepacia infection also led to the accumulation of inflammatory cells and exudates in the alveolar spaces of ΔF508 lungs (Fig. 6C and left lower part). To determine whether rapamycin treatment rescues the appearance of the ΔF508 lung tissue during B. cepacia infection, mice were treated with rapamycin and infected intra-tracheally with B. cepacia as described above. H&E stained lung sections of rapamycin-treated mice showed that rapamycin treatment drastically reduced the recruitment of inflammatory cells throughout the CF lung, prevented peribronchiolar and perivascular hemorrhage and preserved the alveolar space of infected ΔF508 mice (Fig. 6C and right middle and lower parts). Therefore, rapamycin treatment decreases inflammation and thus rescues the lung tissue in a mouse model of CF following B. cepacia infection.
Rapamycin treatment decreases B. cepacia recovery from ΔF508 macrophages by enhancing their delivery to lysosomes.
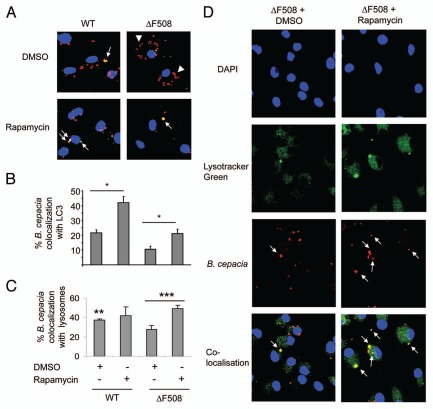
Rapamycin treatment significantly decreased the recovery of B. cepacia from ΔF508 mice and their derived macrophages, yet the mechanism is unclear. It is possible that rapamycin-mediated induction of autophagy enhanced the maturation and the fusion of the B. cepacia-enclosing vacuole with the lysosomal compartment. To explore the trafficking of B. cepacia within macrophages, the colocalization of mRFP-expressing B. cepacia with the autophagy marker LC3 was determined in the presence of rapamycin. Rapamycin increased the number of B. cepacia-containing vacuole that acquired LC3 in both WT and ΔF508 macrophages (Fig. 7A; white arrows and B). Since rapamycin promoted the acquisition of LC3, we examined if it enables the delivery of B. cepacia to the lysosome. Green lysotracker, which accumulates in lysosomes, was added to rapamycin or DMSO-treated WT and ΔF508 macrophages prior to infection with mRFP-expressing B. cepacia. Almost 40% and 20% of B. cepacia localized with the lysotracker in DMSO-treated WT and ΔF508 macrophages respectively within 2 h infection indicating that WT macrophages are more efficient in delivering B. cepacia to the lysosomes (Figs. 7A, B and S7). Rapamycin treatment slightly increased the number of B. cepacia that trafficked to the lysosome in WT macrophages. Nevertheless, rapamycin treatment significantly increased the delivery of B. cepacia to the lysosomes in ΔF508 macrophages up to 50% within 2 h (Figs. 7C, D and S7). Therefore, stimulation of autophagy by rapamycin drives the maturation of the B. cepacia-containing vacuole enabling it to fuse with the lysosome and thereby improving the clearance of the bacterium from ΔF508 macrophages.
Figure 7.
Treatment with rapamycin increases B. cepacia colocalization with LC3/Atg8 and with lysosomes. Bone marrow derived macrophages (BMDM) from wild-type (WT) and BMDM harboring ΔF508 mutation (ΔF508) were treated with rapamycin or DMSO for 1 h before infection, and then macrophages were infected with B. cepacia for 2 h. (A) confocal microscopy demonstrates nuclei stained with DAPI, LC3 stained green and B. cepacia expressing monomeric red florescent protein (mRFP). White arrows indicate the sites of colocalization with of B. cepacia with LC3 with or without rapamycin. White arrows indicate colocalization, white arrow heads indicate B. cepacia. (B) scoring of the percentage of colocalization of B. cepacia with LC3 with or without rapamycin treatment. (C) scoring of the percentage of colocalization of B. cepacia with Lystracker Green with or without rapamycin. (D) confocal microscopy showing nuclei stained with DAPI, lysosomes with Lysotracker Green and mRFP-expressing B. cepacia. the site of colocalization of B. cepacia with Lysotracker Green in ΔF508 macrophages upon rapamycin (ΔF508 + rapamycin) or DMSO (ΔF508 + DMSO) are indicated with white arrows. Data are representative of 3 independent experiments and presented as the means ± SD. Asterisks in (B and c) indicate significant differences from the DMSO-treated cells (*p < 0.05; **p < 0.01; ***p < 0.001).
Discussion
CF is the most common life-shortening inherited disease among the Caucasian population owing in part to the exaggerated pulmonary inflammatory response in the lungs of CF patients upon infection especially with B. cepacia.59,67,68 B. cepacia is transmissible among CF patients and impossible to eradicate due to its inherent antibiotic resistance. Why CF patients are specifically susceptible to B. cepacia is unknown. Here we demonstrate that B. cepacia has a survival advantage in ΔF508 macrophages as the number of B. cepacia associated with ΔF508 macrophages is strikingly higher than those associated with WT macrophages. The results raise the possibility that B. cepacia uses the macrophages as a sanctuary to evade host defense.
The nature of the B. cepacia-containing vacuole and the mechanism halting its fusion with the lysosome were unclear. Here we show that in WT macrophages, a fraction of intracellular B. cepacia resides in multilayered LC3-labeled compartments consistent with autophagosomes.69 A percentage of the B. cepaciacontaining autophagosomes fuses with the lysosome. In contrast, in ΔF508 macrophages, LC3 is not detected around the B. cepacia vacuole, which rarely fuses with the lysosome (Fig. 7C). Electron microscopy data (Fig. 2D) showed that in WT macrophages, a fraction of intracellular B. cepacia resides in multilayered vacuoles that resemble the autophagosomes while in ΔF508 macrophages, most bacteria remained inside single-membrane vacuoles.
Autophagy stimulation with rapamycin in ΔF508 macrophages enhances the delivery of B. cepacia to the lysosomes. Interestingly, the small GTP-binding protein Rab7 is inactivated by B. cepacia.49 Rab7 is required for the final maturation of the late autophagic vacuole.70 Thus, this observation is consistent with the lack of autophagy markers on the B. cepacia vacuole in ΔF508 macrophages.
Our data suggest that the inherently defective autophagy activity in ΔF508 cells11,12 in concert with the downregulation of major autophagy genes and inactivation of Rab7 by B. cepacia contribute to the persistence of B. cepacia in ΔF508 macrophages.
Why CF patients are more prone to B. cepacia infection and not to other autophagy-interacting pathogenic bacteria such as Salmonella is unknown. It seems that despite its compromised activity in ΔF508 macrophages, autophagy is still effective in clearing other autophagy-interacting bacteria. One possibility is that other intracellular bacteria like Salmonella do not decrease the expression levels of autophagy molecules. Thus, the down-regulation of autophagy gene expression appears to give B. cepacia a survival advantage in ΔF508 macrophages. Suppression of autophagy has been employed by other pathogens such as Francisella or Listeria.71–73 Yet, it seems that their strong autophagy-suppression capacity gives these pathogens a survival advantage even in WT cells, unlike B. cepacia.
During B. cepacia infection, caspase-1 is activated and pro-IL-1β is cleaved and released from infected macrophages.74 More IL-1β is produced from ΔF508 macrophages than WT macrophages during B. cepacia infection. This could be due to increased B. cepacia burden in ΔF508 macrophages. It is also possible that IL-1β release is higher in ΔF508 cells due to defective autophagy irrespective of the bacterial burden as suggested by a study demonstrating that autophagy regulates IL-1β secretion in response to lipopolysaccharide (LPS) by targeting pro-IL-1β for degradation.75,76 It is also plausible that both factors contribute to excess IL-1β production in ΔF508 macrophages infected with B. cepacia.
We found that rapamycin treatment reduces the production of inflammatory cytokines in vitro. H&E stained sections of infected lungs showed few focal regions of inflammation within WT infected lungs with the preservation of some healthy lung tissue. In contrast, stained sections of ΔF508 lungs showed the accumulation of inflammatory cells in the peribronchiolar and perivascular areas. Alveolar spaces were filled with inflammatory cells and with exudates. Treatment of WT mice with rapamycin pre- and post-infection improved the preservation of healthy lung tissue. The effect of rapamycin treatment on CF lungs was most impressive because the lungs of CF mice treated with rapamycin were spared from the diffuse and intense inflammatory infiltrate observed in mice that did not receive rapamycin. Recent work has showed that human and mouse CF airway epithelia are autophagy deficient and rescued by cystamine, an autophagyinducing molecule, as it favored the clearance of CFTR aggregates.11,12 Therefore, the recognition of the role of autophagy in B. cepacia infection will lead to the development of a novel class of therapeutic agents that will clear CF aggregates and B. cepacia infection simultaneously.77,78 Thus, our findings have the potential for clinical application in CF patients who currently have limited options for treatment of B. cepacia infection and its associated deleterious inflammation.
Materials and Methods
Bone-marrow-derived macrophages.
All animal experiments were performed according to protocols approved by the Animal Care Use Committee of the Ohio State University College of Medicine. Wild-type (WT) C57BL/6, ΔF508 mice were obtained from Case Western University and housed in the OSU vivarium. Bone marrow-derived macrophages (BMDMs) were isolated from the femurs of 6- to 12-wk-old mice and were cultured in IMDM (GIBCO, 12440) containing 10% heat-inactivated FBS (GIBCO, 16000), 20% L cell-conditioned medium, 100 U/ml penicillin and 100 mg/ml streptomycin (GIBCO, 15140) at 37°C in a humidified atmosphere containing 5% CO2. Macrophages were infected with B. cepacia K56-2 or the corresponding gentamicin sensitive strain MHK1 at a multiplicity of infection (MOI) of 10.
Bacterial strains and culture.
Burkholderia cepacia strain K56-2 was isolated from a CF patient. All bacterial strains were grown in Luria-Bertani (LB) broth at 37°C overnight with high amplitude shaking. The B. cepacia MHK1 strain has a mutation in an antibiotic efflux pump that confers gentamicin sensitivity but does not alter the trafficking of the mutant in macrophages.79 To kill extracellular bacteria, Iscove's media (GIBCO, 12440) containing 10% heat-inactivated FBS (GIBCO, 16000) containing 50 µg/ml gentamicin (GIBCO, 3564) was added for 30 min as previously described in reference 79. To enumerate intracellular bacteria, infected macrophages were lysed using ice cold PBS (GIBCO, 14190) and physical disruption. Macrophages from monolayers were scrapped and pipetted repeatedly against the walls and bottom of the well. Recovered bacteria were quantified by plating serial dilutions on LB agar plates and counting colonies using the Acolyte Colony Counter, 5710/SYN.
Mouse in vivo infection.
WT C57BL/6 and ΔF508 mice were infected intra-tracheally with 10 x 106 WT bacteria with rapamycin (Sigma-Aldrich, R0395) or DMSO (Sigma-Aldrich, D2650) treatment (n = 3). Rapamycin was used in vivo at 4 mg/kg by intra-peritoneal injections. Mice were pretreated with two doses of rapamycin for 2 d (24 h interval), and then infected with B. cepacia 2 h after the second dose of rapamycin. On the third day, mice treated with a final dose of rapamycin. The mice were sacrificed 2 h later, and the number of bacteria in the lungs was determined at second day post-infection.14 All animal experiments were performed according to animal protocols approved by the Animal Care Use Committee of the Ohio State University College of Medicine.
In vitro rapamycin treatment.
Rapamycin (Sigma-Aldrich, R0395) was dissolved in DMSO (Sigma-Aldrich, D2650) at 1 mg/ml. Rapamycin was used at concentration 5 µg/ml, DMSO alone was used as a diluent control.
Immunoblotting.
Macrophages were stimulated with B. cepacia K56-2 (MHK1) and the culture supernatant was removed. The cells were washed twice with PBS (GIBCO, 14190) and lysed in lysis buffer solution supplemented with a protease inhibitor mixture (Roche Applied Science, 10-519-978-001). The protein concentration was adjusted to 40 µg/ml. Proteins were separated by sodium dodecyl sulfate-15% PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad Laboratories, 1620117). Membranes were immunoblotted for pro-Interleukin-1β (IL-1β) antibody (kindly provided by Dr. Mark Wewers, Ohio State University) and Atg8 (LC3) (Sigma-Aldrich, L8918) protein bands were detected with secondary antibodies conjugated to horseradish peroxidase followed by enhanced chemiluminescence reagents (Amersham ECL protein gel blotting detection reagents GE Health Care-Life Sciences, RPN2106).
Enzyme-linked immuno sorbent assay (ELISA).
Macrophages were infected with B. cepacia K56-2 (MH1K) for different time points 6, 8, 24 h. Then, culture supernatants were collected, centrifuged and stored at -20°C until assayed for cytokine content. The amounts of IL-1β, IL-6 and TNFα in the supernatant were determined by specific sandwich ELISA following the manufacturer's protocol (R&D System Inc., DY201, DY406, DY410 respectively) and as previously described in reference 80.
siRNA knockdown of LC3.
siRNA treatment was performed using siRNA against LC3 (Dharmacon, J-040989-09): CUA AUA AAG GCA CAA CGA A, GGA UAU AGC UCU AAG CCG G, CAU CCU AAG UUG CCA AUA A, ACU AUG GUG CGA UCA GUA A. siRNA was nucleofected into primary murine macrophages using Lonza Nucleofection kit (VPA-1009) and Amaxa equipment (AAD-10015) as we described previously in references 81 and 82.
Real time PCR.
Total RNA was isolated from cells were lysed in Trizol (Invitrogen Life Technologies, 15596-026) and submitted to SA Biosciences for autophagy array study. Gene expression was calculated as relative copy numbers (RCN), as described previously in references 81 and 83. Briefly, Ct values of every target gene were subtracted from the average Ct of five housekeeping genes, present on the autophagy array (Gusb, Hprt1, Hsp90ab1, Gapdh, Actb) and the resulted ΔCt was used in the equation: RCN = (2-ΔCt) × 100. RCN for every gene represents its expression as number of copies relative to the 100 copies of average housekeeping genes.81,83
Histopathological analysis.
Lungs were removed en bloc and fixed in (10% Formalin) at room temperature for 24 h then formalin was replaced by PBS (GIBCO, 14190), and processed for paraffin embedding. Formalin-preserved sections of the lungs we processed and embedded in paraffin by standard techniques. Sections of 5 µm thick were stained with hematoxylin and eosin (H&E) and examined. Longitudinal sections of 5 µm taken at regular intervals were obtained using a microtome from the proximal, medial and distal lung regions.
Confocal microscopy.
Immunofluorescence microscopy experiments were performed as previously described in reference 84. B. cepacia expressing monomeric Red Fluorescent Protein (mRFP) was used. Localization of markers on B. cepacia phagosomes was performed as previously described in references 14 and 63. Antibodies used were rabbit anti-Atg8/LC3 (Abgent, AP1805a) followed by fluorescent secondary antibodies (Molecular Probes, A11008). Nuclei were stained with the nucleic acid dye 4′,6′-diamino-2-phenylindole and lysosomes were stained green with Lysotracker Green (Invitrogen, L-7526) as previously described in references 14, 63, 80 and 85. In each experiment at least 100 bacteria were scored. Experiments were performed at least three times. Samples were analyzed with (Olympus FV 1000 Spectral Confocal) at the Ohio State University Microscopy Core Facility.
Transmission electron microscopy.
WT and ΔF508 primary murine macrophages cultured on Permanox (Lab-Tek) chamber slides were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer with 0.1 M sucrose. The cells were postfixed with 1% osmium tetroxide in phosphate buffer and then en bloc stained with 2% uranyl acetate in 10% ethanol, dehydrated in a graded series of ethanol and embedded in Eponate 12 epoxy resin (Ted Pella Inc., 18012). Ultrathin sections were cut on a Leica EM UC6 ultramicrotome (Leica microsystems, EM FC7), collected on copper grids, and then stained with Lead citrate and rranyl acetate. Images were acquired with an FEI Technai G2 Spirit transmission electron microscope (FEI), and Macrofire (Optronics) digital camera and AMT image capture software.
Statistical analysis.
All experiments were performed at least three independent times and yielded similar results. Comparisons of groups for statistical difference were done using Student's two-tailed t test. p value ≤ 0.05 was considered significant.
Acknowledgements
Images used in this article were generated at the Campus Microscopy and Imaging Facility, the Ohio State University. We thank Olga Gavrilina for raw RCN calculations and statistical analysis. Basant A. Abdulrahman and Dalia H.A. Abdelaziz are supported by a doctoral fellowship from the Egyptian Bureau of Education. M. Valvano's research is supported by grants from Cystic Fibrosis Canada. M. Valvano holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis and a Zeller's Senior Researcher Award from Cystic Fibrosis Canada. Studies in Dr. Amal Amer's laboratory are supported by grants R01HL094586 and R21Al083871 and the American Lung Association.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Our protocol number 2007A0070 has been approved by the OSU Institutional Animal Care and Use Committee (IACUC). All efforts were made to minimize suffering.
Supplementary Material
References
- 1.Witko-Sarsat V, Sermet-Gaudelus I, Lenoir G, Descamps-Latscha B. Inflammation and CFTR: might neutrophils be the key in cystic fibrosis? Mediators Inflamm. 1999;8:7–11. doi: 10.1080/09629359990658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knorre A, Wagner M, Schaefer HE, Colledge WH, Pahl HL. DeltaF508-CFTR causes constitutive NFkappaB activation through an ER-overload response in cystic fibrosis lungs. Biol Chem. 2002;383:271–282. doi: 10.1515/BC.2002.029. [DOI] [PubMed] [Google Scholar]
- 3.Perez A, Issler AC, Cotton CU, Kelley TJ, Verkman AS, Davis PB. CFTR inhibition mimics the cystic fibrosis inflammatory profile. Am J Physiol Lung Cell Mol Physiol. 2007;292:383–395. doi: 10.1152/ajplung.00403.2005. [DOI] [PubMed] [Google Scholar]
- 4.Konstan MW, Berger M. Current understanding of the inflammatory process in cystic fibrosis: onset and etiology. Pediatr Pulmonol. 1997;24:137–142. doi: 10.1002/(SICI)1099-0496(199708)24:2<137::AIDPPUL13>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 5.O'Malley CA. Infection control in cystic fibrosis: cohorting, cross-contamination and the respiratory therapist. Respir Care. 2009;54:641–657. doi: 10.4187/aarc0446. [DOI] [PubMed] [Google Scholar]
- 6.Webb AK. The difficulties of treating infection in adults with cystic fibrosis. Monaldi Arch Chest Dis. 1993;48:657–661. [PubMed] [Google Scholar]
- 7.Murphy BS, Bush HM, Sundareshan V, Davis C, Hagadone J, Cory TJ, et al. Characterization of macrophage activation states in patients with cystic fibrosis. J Cyst Fibros. 2010;9:314–322. doi: 10.1016/j.jcf.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Armstrong DS, Grimwood K, Carlin JB, Carzino R, Gutièrrez JP, Hull J, et al. Lower airway inflammation in infants and young children with cystic fibrosis. Am J Respir Crit Care Med. 1997;156:1197–1204. doi: 10.1164/ajrccm.156.4.96-11058. [DOI] [PubMed] [Google Scholar]
- 9.Khan TZ, Wagener JS, Bost T, Martinez J, Accurso FJ, Riches DW. Early pulmonary inflammation in infants with cystic fibrosis. Am J Respir Crit Care Med. 1995;151:1075–1082. doi: 10.1164/ajrccm/151.4.1075. [DOI] [PubMed] [Google Scholar]
- 10.Rajan S, Saiman L. Pulmonary infections in patients with cystic fibrosis. Semin Respir Infect. 2002;17:47–56. doi: 10.1053/srin.2002.31690. [DOI] [PubMed] [Google Scholar]
- 11.Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina D, Settembre C, et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat Cell Biol. 2010;12:863–875. doi: 10.1038/ncb2090. [DOI] [PubMed] [Google Scholar]
- 12.Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina DL, Settembre C, et al. Cystic fibrosis: a disorder with defective autophagy. Autophagy. 2011;7:104–106. doi: 10.4161/auto.7.1.13987. [DOI] [PubMed] [Google Scholar]
- 13.Deretic V. Autophagy in infection. Curr Opin Cell Biol. 2010;22:252–262. doi: 10.1016/j.ceb.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amer AO, Byrne BG, Swanson MS. Macrophages rapidly transfer pathogens from lipid raft vacuoles to autophagosomes. Autophagy. 2005;1:53–58. doi: 10.4161/auto.1.1.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brest P, Corcelle EA, Cesaro A, Chargui A, Belaïd A, Klionsky DJ, et al. Autophagy and Crohn's disease: at the crossroads of infection, inflammation, immunity and cancer. Curr Mol Med. 2010;10:486–502. doi: 10.2174/156652410791608252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klionsky DJ. The molecular machinery of autophagy and its role in physiology and disease. Semin Cell Dev Biol. 2010;21:663. doi: 10.1016/j.semcdb.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- 19.Vergne I, Deretic V. The role of PI3P phosphatases in the regulation of autophagy. FEBS Lett. 2010;584:1313–1318. doi: 10.1016/j.febslet.2010.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 22.Wang CW, Klionsky DJ. The molecular mechanism of autophagy. Mol Med. 2003;9:65–76. [PMC free article] [PubMed] [Google Scholar]
- 23.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 24.Mizushima N, Yoshimori T, Ohsumi Y. Role of the Apg12 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2003;35:553–561. doi: 10.1016/S1357-2725(02)00343-6. [DOI] [PubMed] [Google Scholar]
- 25.Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–448. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 26.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 27.Sajjan US, Yang JH, Hershenson MB, LiPuma JJ. Intracellular trafficking and replication of Burkholderia cenocepacia in human cystic fibrosis airway epithelial cells. Cell Microbiol. 2006;8:1456–1466. doi: 10.1111/j.1462-5822.2006.00724.x. [DOI] [PubMed] [Google Scholar]
- 28.Zeng X, Kinsella TJ. Mammalian target of rapamycin and S6 kinase 1 positively regulate 6-thioguanine-induced autophagy. Cancer Res. 2008;68:2384–2390. doi: 10.1158/0008-5472.CAN-07-6163. [DOI] [PubMed] [Google Scholar]
- 29.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 30.Orvedahl A, Levine B. Eating the enemy within: autophagy in infectious diseases. Cell Death Differ. 2009;16:57–69. doi: 10.1038/cdd.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yano T, Kurata S. Induction of autophagy via innate bacterial recognition. Autophagy. 2008;4:958–960. doi: 10.4161/auto.6802. [DOI] [PubMed] [Google Scholar]
- 32.Py BF, Lipinski MM, Yuan J. Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy. 2007;3:117–125. doi: 10.4161/auto.3618. [DOI] [PubMed] [Google Scholar]
- 33.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the 29 Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 34.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 35.Zhou D, Spector SA. Human immunodeficiency virus type-1 infection inhibits autophagy. AIDS. 2008;22:695–699. doi: 10.1097/QAD.0b013e3282f4a836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 37.Ledson MJ, Gallagher MJ, Jackson M, Hart CA, Walshaw MJ. Outcome of Burkholderia cepacia colonisation in an adult cystic fibrosis centre. Thorax. 2002;57:142–145. doi: 10.1136/thorax.57.2.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCloskey M, McCaughan J, Redmond AO, Elborn JS. Clinical outcome after acquisition of Burkholderia cepacia in patients with cystic fibrosis. Ir J Med Sci. 2001;170:28–31. doi: 10.1007/BF03167716. [DOI] [PubMed] [Google Scholar]
- 39.Valvano MA, Keith KE, Cardona ST. Survival and persistence of opportunistic Burkholderia species in host cells. Curr Opin Microbiol. 2005;8:99–105. doi: 10.1016/j.mib.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Vergunst AC, Meijer AH, Renshaw SA, O'Callaghan D. Burkholderia cenocepacia creates an intramacrophage replication niche in zebrafish embryos, followed by bacterial dissemination and establishment of 30 systemic infection. Infect Immun. 2010;78:1495–1508. doi: 10.1128/IAI.00743-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marolda CL, Hauroder B, John MA, Michel R, Valvano MA. Intracellular survival and saprophytic growth of isolates from the Burkholderia cepacia complex in free-living amoebae. Microbiology. 1999;145:1509–1517. doi: 10.1099/13500872-145-7-1509. [DOI] [PubMed] [Google Scholar]
- 42.Hunt TA, Kooi C, Sokol PA, Valvano MA. Identification of Burkholderia cenocepacia genes required for bacterial survival in vivo. Infect Immun. 2004;72:4010–4022. doi: 10.1128/IAI.72.7.4010-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamothe J, Huynh KK, Grinstein S, Valvano MA. Intracellular survival of Burkholderia cenocepacia in macrophages is associated with a delay in the maturation of bacteria-containing vacuoles. Cell Microbiol. 2007;9:40–53. doi: 10.1111/j.1462-5822.2006.00766.x. [DOI] [PubMed] [Google Scholar]
- 44.Saini LS, Galsworthy SB, John MA, Valvano MA. Intracellular survival of Burkholderia cepacia complex isolates in the presence of macrophage cell activation. Microbiology. 1999;145:3465–3475. doi: 10.1099/00221287-145-12-3465. [DOI] [PubMed] [Google Scholar]
- 45.LiPuma JJ. Burkholderia and emerging pathogens in cystic fibrosis. Semin Respir Crit Care Med. 2003;24:681–692. doi: 10.1055/s-2004-815664. [DOI] [PubMed] [Google Scholar]
- 46.Sajjan US, Hershenson MB, Forstner JF, LiPuma JJ. Burkholderia cenocepacia ET12 strain activates TNFR1 signalling in cystic fibrosis airway epithelial cells. Cell Microbiol. 2008;10:188–201. doi: 10.1111/j.1462-5822.2007.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamothe J, Valvano MA. Burkholderia cenocepaciainduced delay of acidification and phagolysosomal fusion in cystic fibrosis transmembrane conductance regulator (CFTR)-defective macrophages. Microbiology. 2008;154:3825–3834. doi: 10.1099/mic.0.2008/023200-0. [DOI] [PubMed] [Google Scholar]
- 48.Keith KE, Hynes DW, Sholdice JE, Valvano MA. Delayed association of the NADPH oxidase complex with macrophage vacuoles containing the opportunistic pathogen Burkholderia cenocepacia. Microbiology. 2009;155:1004–1015. doi: 10.1099/mic.0.026781-0. [DOI] [PubMed] [Google Scholar]
- 49.Huynh KK, Plumb JD, Downey GP, Valvano MA, Grinstein S. Inactivation of macrophage Rab7 by Burkholderia cenocepacia. J Innate Immun. 2010;2:522–533. doi: 10.1159/000319864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Armstrong DS, Hook SM, Jamsen KM, Nixon GM, Carzino R, Carlin JB, et al. Lower airway inflammation in infants with cystic fibrosis detected by newborn screening. Pediatr Pulmonol. 2005;40:500–510. doi: 10.1002/ppul.20294. [DOI] [PubMed] [Google Scholar]
- 51.Rosenfeld M, Gibson RL, McNamara S, Emerson J, Burns JL, Castile R, et al. Early pulmonary infection, inflammation and clinical outcomes in infants with cystic fibrosis. Pediatr Pulmonol. 2001;32:356–366. doi: 10.1002/ppul.1144. [DOI] [PubMed] [Google Scholar]
- 52.Reiniger N, Lee MM, Coleman FT, Ray C, Golan DE, Pier GB. Resistance to Pseudomonas aeruginosa chronic lung infection requires cystic fibrosis transmembrane conductance regulator-modulated Interleukin-1 (IL-1) release and signaling through the IL-1 receptor. Infect Immun. 2007;75:1598–1608. doi: 10.1128/IAI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levy H, Murphy A, Zou F, Gerard C, Klanderman B, Schuemann B, et al. IL1B polymorphisms modulate cystic fibrosis lung disease. Pediatr Pulmonol. 2009;44:580–593. doi: 10.1002/ppul.21026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corvol H, Fitting C, Chadelat K, Jacquot J, Tabary O, Boule M, et al. Distinct cytokine production by lung and blood neutrophils from children with cystic fibrosis. Am J Physiol Lung Cell Mol Physiol. 2003;284:997–1003. doi: 10.1152/ajplung.00156.2002. [DOI] [PubMed] [Google Scholar]
- 55.Berger M. Inflammatory mediators in cystic fibrosis lung disease. Allergy Asthma Proc. 2002;23:19–25. [PubMed] [Google Scholar]
- 56.Bonfield TL, Panuska JR, Konstan MW, Hilliard KA, Hilliard JB, Ghnaim H, et al. Inflammatory cytokines in cystic fibrosis lungs. Am J Respir Crit Care Med. 1995;152:2111–2118. doi: 10.1164/ajrccm.152.6.8520783. [DOI] [PubMed] [Google Scholar]
- 57.Greally P, Hussain MJ, Vergani D, Price JF. Serum interleukin-1alpha and soluble interleukin-2 receptor concentrations in cystic fibrosis. Arch Dis Child. 1993;68:785–787. doi: 10.1136/adc.68.6.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kronborg G, Hansen MB, Svenson M, Fomsgaard A, Høiby N, Bendtzen K. Cytokines in sputum and serum from patients with cystic fibrosis and chronic Pseudomonas aeruginosa infection as markers of destructive inflammation in the lungs. Pediatr Pulmonol. 1993;15:292–297. doi: 10.1002/ppul.1950150506. [DOI] [PubMed] [Google Scholar]
- 59.Kotrange S, Kopp B, Akhter A, Abdelaziz D, Abu Khweek A, Caution K, et al. Burkholderia cenocepacia O polysaccharide chain contributes to caspase-1-dependent IL-1{beta} production in macrophages. J Leukoc Biol. 2011;89:481–488. doi: 10.1189/jlb.0910513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du K, Sharma M, Lukacs GL. The DeltaF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat Struct Mol Biol. 2005;12:17–25. doi: 10.1038/nsmb882. [DOI] [PubMed] [Google Scholar]
- 61.Sugarman EA, Rohlfs EM, Silverman LM, Allitto BA. CFTR mutation distribution among USHispanic and African American individuals: evaluation in cystic fibrosis patient and carrier screening populations. Genet Med. 2004;6:392–399. doi: 10.1097/01.GIM.0000139503.22088.66. [DOI] [PubMed] [Google Scholar]
- 62.Roqué M, Godoy CP, Castellanos M, Pusiol E, Mayorga LS. Population screening of F508del (DeltaF508), the most frequent mutation in the CFTR gene associated with cystic fibrosis in Argentina. Hum Mutat. 2001;18:167. doi: 10.1002/humu.1173. [DOI] [PubMed] [Google Scholar]
- 63.Amer AO, Swanson MS. Autophagy is an immediate macrophage response to Legionella pneumophila. Cell Microbiol. 2005;7:765–778. doi: 10.1111/j.1462-5822.2005.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu J, Parkhitko AA, Henske EP. Mammalian target of rapamycin signaling and autophagy: roles in lymphangioleiomyomatosis therapy. Proc Am Thorac Soc. 2010;7:48–53. doi: 10.1513/pats.200909-104JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shigemitsu K, Tsujishita Y, Hara K, Nanahoshi M, Avruch J, Yonezawa K. Regulation of translational effectors by amino acid and mammalian target of rapamycin signaling pathways. Possible involvement of autophagy in cultured hepatoma cells. J Biol Chem. 1999;274:1058–1065. doi: 10.1074/jbc.274.2.1058. [DOI] [PubMed] [Google Scholar]
- 66.Harris J, Hartman M, Roche C, Zeng SG, O'Shea A, Sharp FA, et al. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem. 2011;286:9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hendry J, Elborn JS, Nixon L, Shale DJ, Webb AK. Cystic fibrosis: inflammatory response to infection with Burkholderia cepacia and Pseudomonas aeruginosa. Eur Respir J. 1999;14:435–438. doi: 10.1034/j.1399-3003.1999.14b32.x. [DOI] [PubMed] [Google Scholar]
- 68.Hendry J, Butler S, Elborn JS, Govan JR, Nelson J, Shale DJ, et al. Antibody response to Burkholderia cepacia in patients with cystic fibrosis colonized with Burkholderia cepacia and Pseudomonas aeruginosa. J Infect. 2000;40:164–170. doi: 10.1053/jinf.1999.0626. [DOI] [PubMed] [Google Scholar]
- 69.Klionsky DJ. Autophagy. Curr Biol. 2005;15:282–283. doi: 10.1016/j.cub.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 70.Noda T, Yoshimori T. Between canonical and antibacterial autophagy: Rab7 is required for GAS-containing autophagosome-like vacuole formation. Autophagy. 2010;6:419–420. doi: 10.4161/auto.6.3.11419. [DOI] [PubMed] [Google Scholar]
- 71.Ogawa M, Yoshikawa Y, Mimuro H, Hain T, Chakraborty T, Sasakawa C. Autophagy targeting of Listeria monocytogenes and the bacterial countermeasure. Autophagy. 2011;7:310–314. doi: 10.4161/auto.7.3.14581. [DOI] [PubMed] [Google Scholar]
- 72.Cremer TJ, Amer A, Tridandapani S, Butchar JP. Francisella tularensis regulates autophagy-related host cell signaling pathways. Autophagy. 2009;5:125–128. doi: 10.4161/auto.5.1.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Butchar JP, Cremer TJ, Clay CD, Gavrilin MA, Wewers MD, Marsh CB, et al. Microarray analysis of human monocytes infected with Francisella tularensis identifies new targets of host response subversion. PLoS ONE. 2008;3:2924. doi: 10.1371/journal.pone.0002924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thornberry NA, Molineaux SM. Interleukin-1 beta converting enzyme: a novel cysteine protease required for IL-1beta production and implicated in programmed cell death. Protein Sci. 1995;4:3–12. doi: 10.1002/pro.5560040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fujishima Y, Nishiumi S, Masuda A, Inoue J, Nguyen NM, Irino Y, et al. Autophagy in the intestinal epithelium reduces endotoxin-induced inflammatory responses by inhibiting NFkappaB activation. Arch Biochem Biophys. 2011;506:223–235. doi: 10.1016/j.abb.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 76.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 77.Leitão JH, Sousa SA, Cunha MV, Salgado MJ, Melo-Cristino J, Barreto MC, et al. Variation of the antimicrobial susceptibility profiles of Burkholderia cepacia complex clonal isolates obtained from chronically infected cystic fibrosis patients: a five-year survey in the major Portuguese treatment center. Eur J Clin Microbiol Infect Dis. 2008;27:1101–1111. doi: 10.1007/s10096008-0552-0. [DOI] [PubMed] [Google Scholar]
- 78.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 79.Hamad MA, Skeldon AM, Valvano MA. Construction of aminoglycoside-sensitive Burkholderia cenocepacia strains for use in studies of intracellular bacteria with the gentamicin protection assay. Appl Environ Microbiol. 2010;76:3170–3176. doi: 10.1128/AEM.03024-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Akhter A, Gavrilin MA, Frantz L, Washington S, Ditty C, Limoli D, et al. Caspase-7 activation by the Nlrc4/Ipaf inflammasome restricts Legionella pneumophila infection. PLoS Pathog. 2009;5:1000361. doi: 10.1371/journal.ppat.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gavrilin MA, Bouakl IJ, Knatz NL, Duncan MD, Hall MW, Gunn JS, et al. Internalization and phagosome escape required for Francisella to induce human monocyte IL-1beta processing and release. Proc Natl Acad Sci USA. 2006;103:141–146. doi: 10.1073/pnas.0504271103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gavrilin MA, Mitra S, Seshadri S, Nateri J, Berhe F, Hall MW, et al. Pyrin critical to macrophage IL-1beta response to Francisella challenge. J Immunol. 2009;182:7982–7989. doi: 10.4049/jimmunol.0803073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fahy RJ, Exline MC, Gavrilin MA, Bhatt NY, Besecker BY, Sarkar A, et al. Inflammasome mRNA expression in human monocytes during early septic shock. Am J Respir Crit Care Med. 2008;177:983–988. doi: 10.1164/rccm.200703-418OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Amer A, Franchi L, Kanneganti TD, Body-Malapel M, Ozören N, Brady G, et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006;281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- 85.Abbott J, Hart A, Morton AM, Dey P, Conway SP, Webb AK. Can health-related quality of life predict survival in adults with cystic fibrosis? Am J Respir Crit Care Med. 2009;179:4–58. doi: 10.1164/rccm.200802-220OC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.