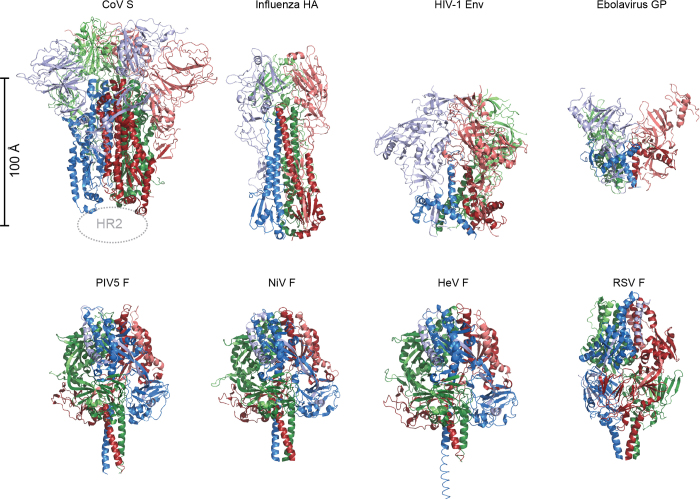
Extended Data Figure 8. Class I viral fusion proteins.
All class I fusion proteins require proteolytic cleavage adjacent to the fusion peptide or loop, and the metastable pre-fusion state is triggered by a series of events that involve pH change or receptor binding. The post-fusion conformations all contain anti-parallel six-helix bundles composed of the HR1 and HR2 from the membrane-proximal subunit. However, there is a great diversity in pre-fusion conformations as shown here. Members of this class that also participate in receptor binding14,15,16,28,53 (top row), including S glycoproteins of coronaviruses, are organized such that their receptor binding subunits sit atop the fusion machinery, and need to be shed in order for membrane fusion to proceed. Paramyxovirus F proteins54,55,56,57 (bottom row) have a different architecture than the capped fusion proteins on the top row. The F proteins all have disulfide bonds between the membrane proximal and membrane distal subunits, and the two subunits remain interconnected throughout the rearrangement process.