A 4.0 Å resolution cryo-electron microscopy structure of the pre-fusion form of the trimeric spike from the human coronavirus HKU1 provides insight into how the spike protein mediates host-cell attachment and membrane fusion.
Supplementary information
The online version of this article (doi:10.1038/nature17200) contains supplementary material, which is available to authorized users.
Subject terms: SARS virus, Cryoelectron microscopy
Coronavirus spike structure
Coronaviruses are responsible for respiratory infections worldwide, many of them mild, but also including severe pneumonia and the recent SARS and MERS outbreaks. The entry of coronaviruses into cells is mediated by the virus glycoprotein spike trimer, which contains the receptor-binding domain, as well as membrane fusion domains. Two papers published in this issue of Nature provide high-resolution (4Å) cryo-electron microscopy structures of pre-fusion coronavirus spike trimers. David Veesler and colleagues studied the trimer from murine hepatitis virus; Andrew Ward and colleagues used the human betacoronavirus HKU1, a cause of mild respiratory disease. The structures reveal mechanistic insights into the viral fusion process and architectural similarities to paramyxovirus F proteins, suggesting that these fusion proteins may have evolved from a distant common ancestor.
Supplementary information
The online version of this article (doi:10.1038/nature17200) contains supplementary material, which is available to authorized users.
Abstract
HKU1 is a human betacoronavirus that causes mild yet prevalent respiratory disease1, and is related to the zoonotic SARS2 and MERS3 betacoronaviruses, which have high fatality rates and pandemic potential. Cell tropism and host range is determined in part by the coronavirus spike (S) protein4, which binds cellular receptors and mediates membrane fusion. As the largest known class I fusion protein, its size and extensive glycosylation have hindered structural studies of the full ectodomain, thus preventing a molecular understanding of its function and limiting development of effective interventions. Here we present the 4.0 Å resolution structure of the trimeric HKU1 S protein determined using single-particle cryo-electron microscopy. In the pre-fusion conformation, the receptor-binding subunits, S1, rest above the fusion-mediating subunits, S2, preventing their conformational rearrangement. Surprisingly, the S1 C-terminal domains are interdigitated and form extensive quaternary interactions that occlude surfaces known in other coronaviruses to bind protein receptors. These features, along with the location of the two protease sites known to be important for coronavirus entry, provide a structural basis to support a model of membrane fusion mediated by progressive S protein destabilization through receptor binding and proteolytic cleavage. These studies should also serve as a foundation for the structure-based design of betacoronavirus vaccine immunogens.
Supplementary information
The online version of this article (doi:10.1038/nature17200) contains supplementary material, which is available to authorized users.
Main
Betacoronavirus S proteins are processed into S1 and S2 subunits by host proteases5. Like other class I viral fusion proteins, the two subunits trimerize and fold into a metastable pre-fusion conformation. The S1 subunit is responsible for receptor binding, while the S2 subunit mediates membrane fusion. Coronaviruses typically possess two domains within S1 capable of binding to host receptors: an amino (N)-terminal domain (NTD) and a carboxy (C)-terminal domain (CTD), with the latter recognizing protein receptors for SARS-CoV and MERS-CoV6,7. Although these individual domains have been structurally characterized, the organization of the complete spike has not yet been determined, preventing a mechanistic understanding of S protein function.
Here, we present the structure of the HKU1 S protein ectodomain determined using cryo-electron microscopy (cryo-EM) to 4.0 Å resolution (Fig. 1a and Extended Data Figs 1 and 2 and Extended Data Table 1). The protein construct contains a C-terminal T4 fibritin trimerization motif and a mutated S1/S2 furin-cleavage site (Extended Data Fig. 3). The S1 subunit adopts an extended conformation with short linkers between domains and sub-domains (Fig. 1b). The S1 NTD (amino acids 14–297) has strong structural and sequence homology to the bovine coronavirus (BCoV) S1 NTD (Extended Data Fig. 4), which recognizes acetylated sialic acids on glycosylated cell-surface receptors8. The glycan-binding site in the BCoV S1 NTD is conserved in the HKU1 S1 NTD and is located at the apex of the trimer, oriented towards target cells. Indeed, HKU1 S1 was recently shown to bind O-acetylated sialic acids on host cells, and these glycans were required for efficient infection of primary human airway epithelial cultures9.
Figure 1. Structure of the HKU1 pre-fusion spike ectodomain.
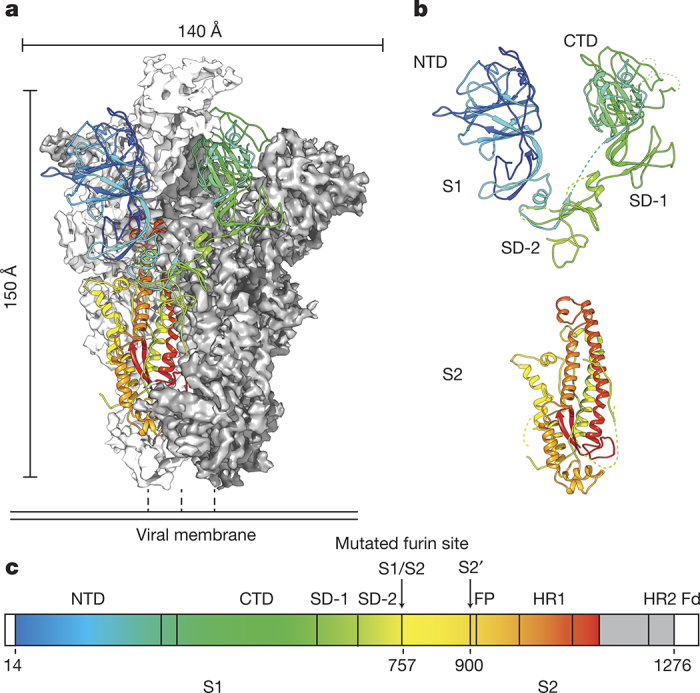
a, A single protomer of the trimeric S protein is shown in cartoon representation coloured as a rainbow from the N to C terminus (blue to red) with the reconstructed EM density of remaining protomers shown in white and grey. b, The S1 subunit is composed of the NTD and CTD as well as two sub-domains (SD-1 and SD-2). The S2 subunit contains the coronavirus fusion machinery and is primarily α-helical. c, Domain architecture of the HKU1 S protein coloured as in a.
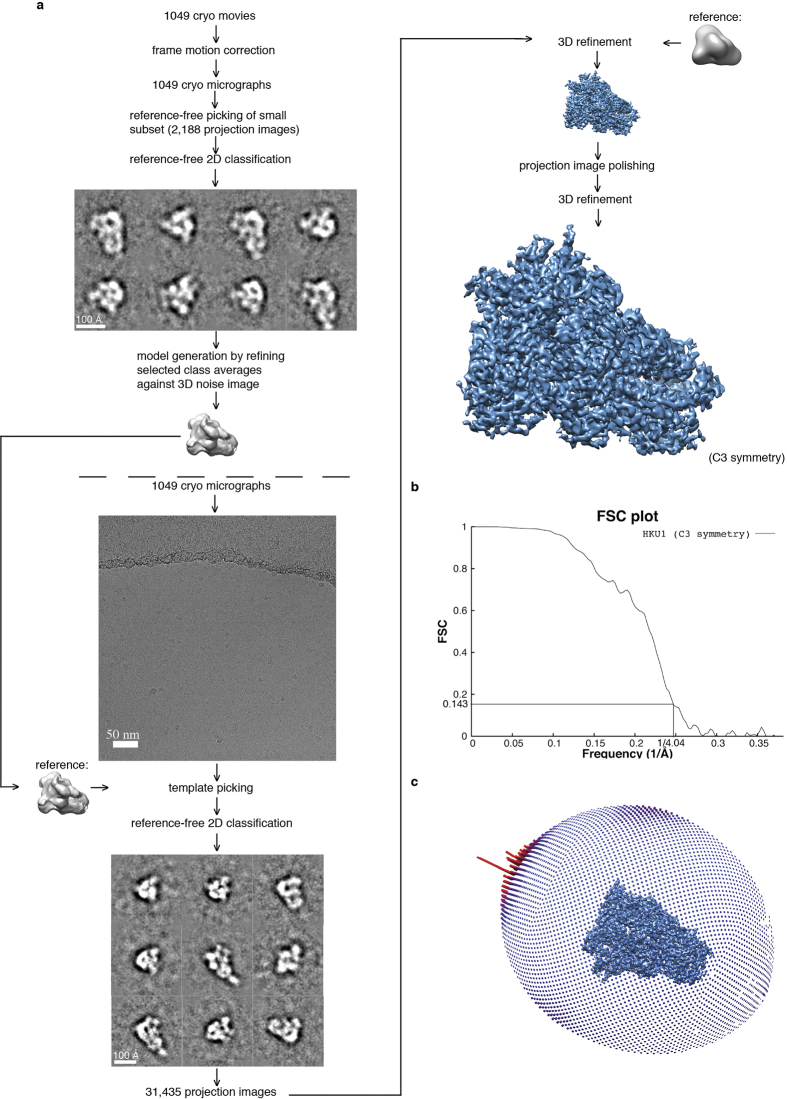
Extended Data Figure 1. Data processing flowchart.
a, Processing resulting in density map of pre-fusion HKU1 spike glycoprotein at 4.04 Å resolution. b, FSC plot illustrating correlation between two volumes refined independently from two distinct half sets of raw data. A final resolution of 4.04 Å is indicated in the plot. c, Angular distribution of raw data within the data set. A slight, but within normal range, over-representation of top views was observed (tall red bars).
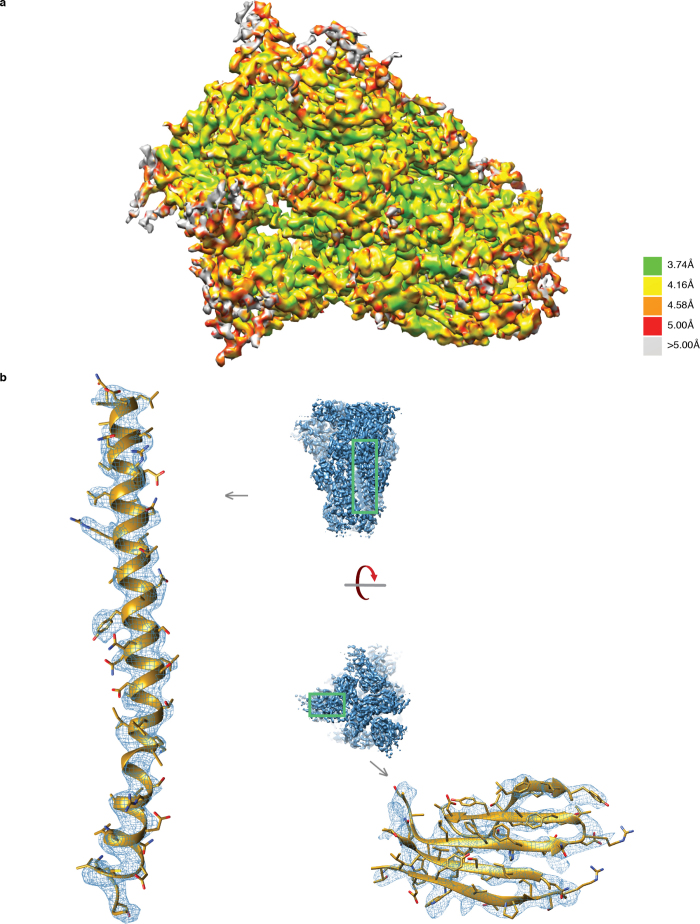
Extended Data Figure 2. Resolution of the pre-fusion HKU1 S density map.
a, Local resolution within the EM density map. Local resolution was calculated using ResMap51 discretizing every 0.25 Å over a range from 2 × voxel size (2.62 Å) to 4 × voxel size (5.24 Å). Resolution significance criterion was set to 0.05. The resolution ranges from 3.74 Å in stable internal secondary structures to greater than 5.00 Å in flexible peripheral loops. b, Close-ups of secondary-structure densities. To the left is displayed the central α-helix of an S2 monomer and to the right is a β-sheet from the NTD domain in an S1 monomer.
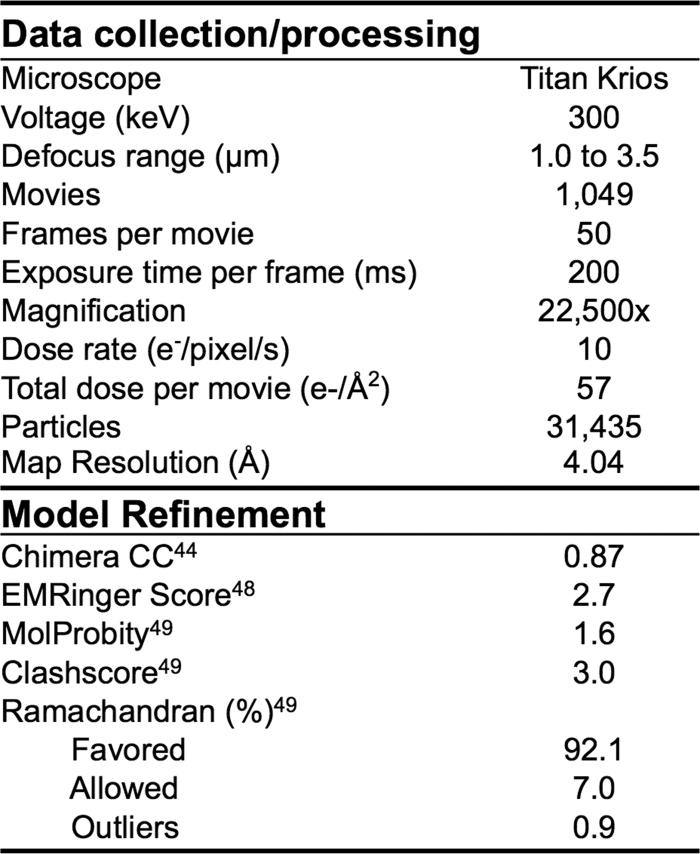
Extended Data Table 1.
CryoEM data collection, processing and refinement metrics
CC = cross correlation
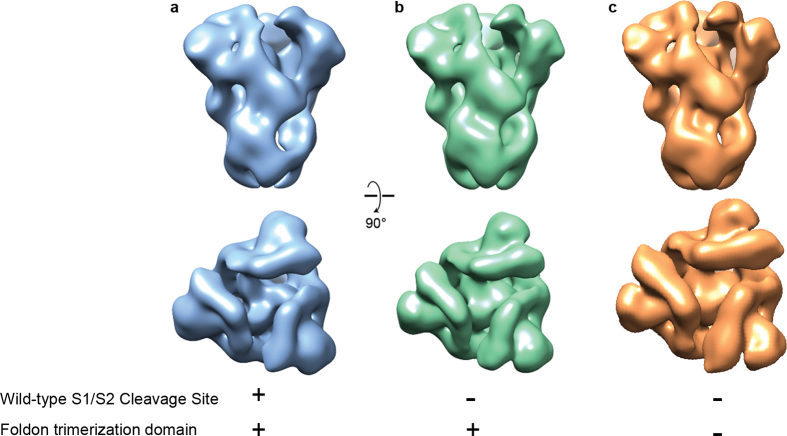
Extended Data Figure 3. Cleavage at the S1/S2 junction does not induce large conformational changes in HKU1 spike.
a, HKU1 spike 1–1249 with an attached foldon domain and wild-type furin-cleavage site was reconstructed using negative-stain electron microscopy. b, HKU1 spike 1–1276 with an attached foldon and a mutated furin-cleavage site reconstructed using negative-stain electron microscopy. c, HKU1 spike 1–1249 without foldon and with mutated furin-cleavage site. Side and top views are shown.
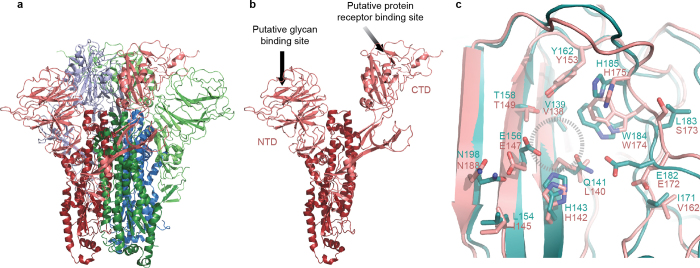
Extended Data Figure 4. Putative glycan binding site of the HKU1 S1 NTD.
a, HKU1 trimeric S and b, an isolated monomer. Putative host glycan-binding and protein-receptor-binding sites are indicated. c, The bovine coronavirus (BCoV) S1 NTD structure from Peng et al.8 (teal) is superposed onto the HKU1 S NTD (pink). Residue side-chains involved in the putative glycan-binding site (dashed circle) are shown as sticks, with oxygen atoms coloured red and nitrogen atoms coloured blue. Note that N198 (BCoV) and N188 (HKU1) are predicted N-linked glycosylation sites.
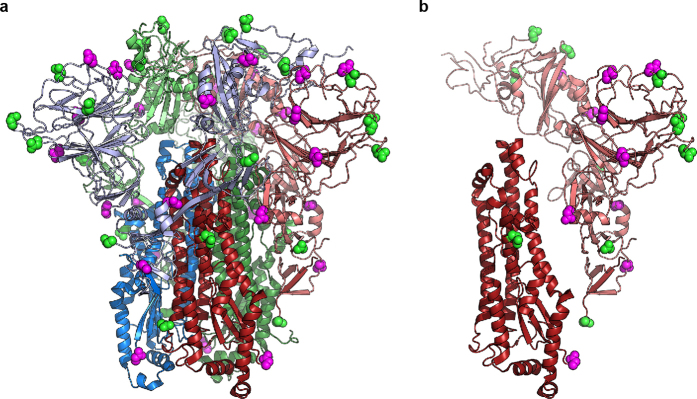
The HKU1 S1 CTD (amino acids 325–605) consists of a structurally conserved core connected to a large, variable loop (HKU1 S amino acids 428–587)10 that is partially disordered (Extended Data Figs 5 and 6). The CTD is located at the trimer apex close to the threefold axis, and the core interacts with the other two S1 CTD cores and with one NTD from an adjacent protomer. The domain swapping between protomers results in a woven appearance when viewed looking down towards the viral membrane (Fig. 2a). Structural alignment of the SARS-CoV and MERS-CoV CTD–receptor complexes11,12 with the HKU1 pre-fusion S protein reveals that the protein-receptor-binding surface of the S1 CTD is buried in the HKU1 S protein trimer and is therefore incapable of making equivalent interactions without some initial breathing and transient exposure of these domains (Fig. 2b). Although a protein receptor has not yet been identified for HKU1, antibodies against the CTD, but not those against the NTD, blocked HKU1 infection of cells13. These data suggest that the S1 CTD is the primary HKU1 receptor-binding site13, whereas the NTD mediates initial attachment via glycan binding.
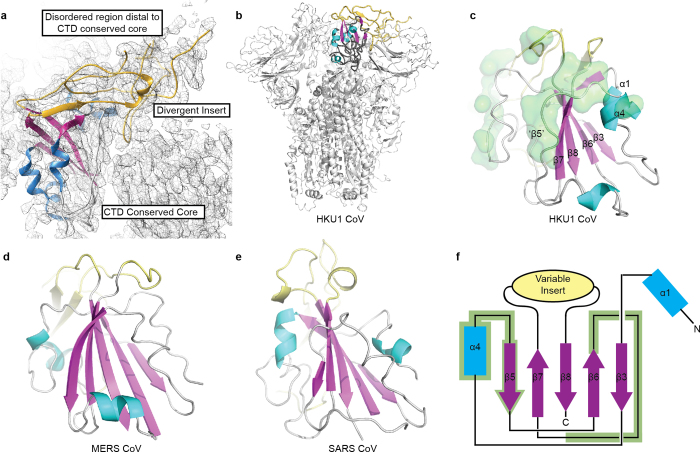
Extended Data Figure 5. Betacoronavirus S proteins possess a conserved structural core in their C-terminal domains.
a, The structurally divergent loop of the S1 CTD is poorly ordered distal to the core CTD domain. The conserved S1 CTD cores10 of b, HKU1-CoV highlighted in the trimeric pre-fusion S, c, HKU1-CoV as an isolated domain, d, MERS-CoV12 and e, SARS-CoV11 are coloured according to secondary structure (β-sheets: pink, α-helices: blue, lacking regular secondary structure: grey) and the insert which differs amongst coronaviruses is coloured yellow. Atoms participating in quaternary interactions with other HKU1 S protomer CTDs are shown in green surface in c. f, The positions of these interacting atoms are mapped on to the conserved core topology. The sheet and helix nomenclature is taken from reference 10.
Extended Data Figure 6. Sequence alignment of human betacoronavirus S proteins.
Sequence alignment of S proteins from HKU1, SARS-CoV and MERS-CoV using Clustal Omega52. Protein features described in the text are indicated: N-terminal domain (NTD), C-terminal domain (CTD) which contains the large variable loop, the S1/S2 and S2′ cleavage sites, fusion peptide (FP), heptad repeats 1 and 2 (HR1, HR2) and transmembrane helix (TM).
Figure 2. Architecture of the HKU1 S1 subunit.
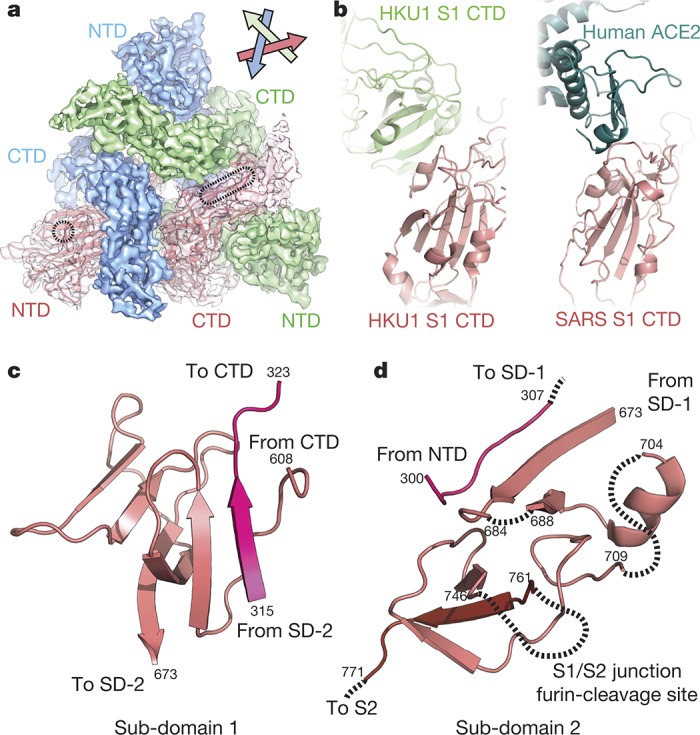
a, EM density corresponding to each S1 protomer is shown. The putative glycan-binding and protein-receptor-binding sites are indicated with dashed shapes on the NTD and CTD, respectively. b, The HKU1 S1 CTD forms quaternary interactions with an adjacent CTD using a surface similar to that used by SARS CTD to bind its receptor, ACE2 (ref. 11). c, Sub-domain 1 is composed of amino acid residues before and after the S1 CTD. d, Sub-domain 2 is composed of S1 sequence C-terminal to the CTD, a short peptide following the NTD, and the N-terminal strand of S2, which follows the S1/S2 furin-cleavage site.
HKU1 S1 also contains two sub-domains (which we term SD-1 and SD-2) that lack significant homology to previously determined structures (Fig. 2c, d). These sub-domains are primarily composed of S1 amino acid sequences following the CTD. However, stretches of amino acids preceding the CTD as well as S2 residues adjacent to the S1/S2 cleavage site also contribute to the sub-domains. This complex folding of elements dispersed throughout the primary sequence may allow receptor-induced conformational changes in the CTD to be transmitted to other parts of the structure.
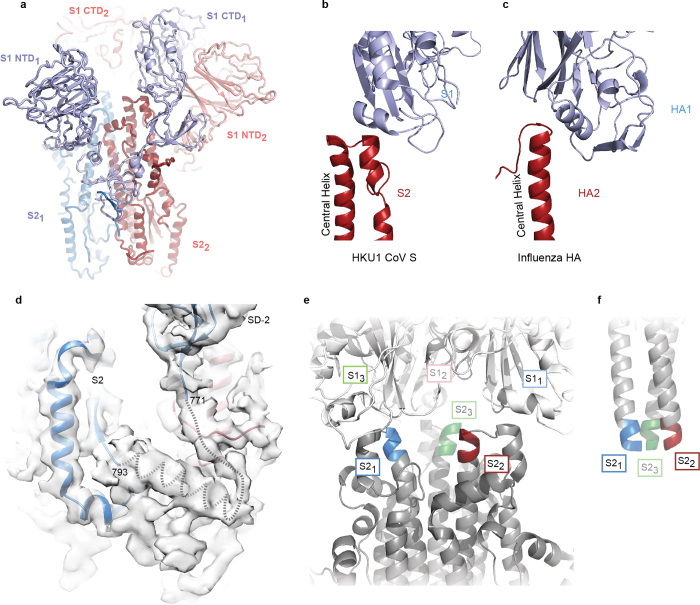
In contrast to other viral fusion proteins such as influenza haemagglutinin (HA)14 or HIV-1 envelope (Env)15,16, the HKU1 S1 subunits are rotated about the trimeric threefold axis with respect to the S2 subunits, causing the S1 subunit from one protomer to sit above the S2 subunit of an adjacent protomer (Extended Data Fig. 7). Similar to HA and Env, a region in the HKU1 S1 CTD (amino acids 371–380) caps the S2 central helix, thereby preventing the fusion machinery from springing into action.
Extended Data Figure 7. S1 sits atop an adjacent protomer’s S2.
a, The HKU1 S1 subunits are rotated about the trimeric threefold axis relative to their corresponding S2 subunits such that the S1 CTD from one protomer caps the S2 central helix from an adjacent protomer (CTD1, blue, caps S22, red). The third protomer of the trimer has been omitted for clarity. b, HKU1 S1 CTD (blue) uses a short helix to cap the central helix and HR1 (red). c, The influenza haemagglutinin HA2 central helix (red) is also capped by a helix in HA1 (blue)14,28. d, The S2 N-terminal β-strand is connected to the remainder of the S2 subunit via a loop and an α-helix (dotted lines). These regions of the EM density are of insufficient quality to confidently build this protein region but enable interpretation of connectivity. e, In the pre-fusion HKU1 S protein, the tops of the central S2 helices (blue, red, green) are splayed outwards from the threefold axis and capped by the S1 CTDs (white). The S1 NTD, SD-1 and SD-2 have been omitted for clarity. f, In the post-fusion six-helix-bundle structure of SARS S22, the corresponding helical regions from (e) form a well-packed three-helix bundle.
Processing of coronavirus S proteins by host proteases plays a critical role in the entry process5. HKU1 S is cleaved by furin into S1 and S2 subunits during protein biosynthesis. Though mutated in the protein construct used here and disordered in the density map, the HKU1 S furin-cleavage site at the S1/S2 junction lies in a loop of SD-2 (Fig. 3 and Extended Data Fig. 6). Furin cleavage would leave a single S2 β-strand participating in the SD-2 β-sheets (Fig. 2d). Coronavirus S proteins also have a secondary cleavage site, termed S2′ (Arg900)5, adjacent to the viral fusion peptide (amino acids 901–918)17 (Fig. 3b and Extended Data Fig. 6). This is similar to the multiple endoproteolytic cleavage events that occur in the fusion proteins of respiratory syncytial virus (RSV) and Ebola virus18,19. Protease cleavage at S2′ likely follows S1/S2 cleavage and may not occur until host-receptor engagement at the plasma membrane or viral endocytosis5.
Figure 3. HKU1 S2 subunit fusion machinery.
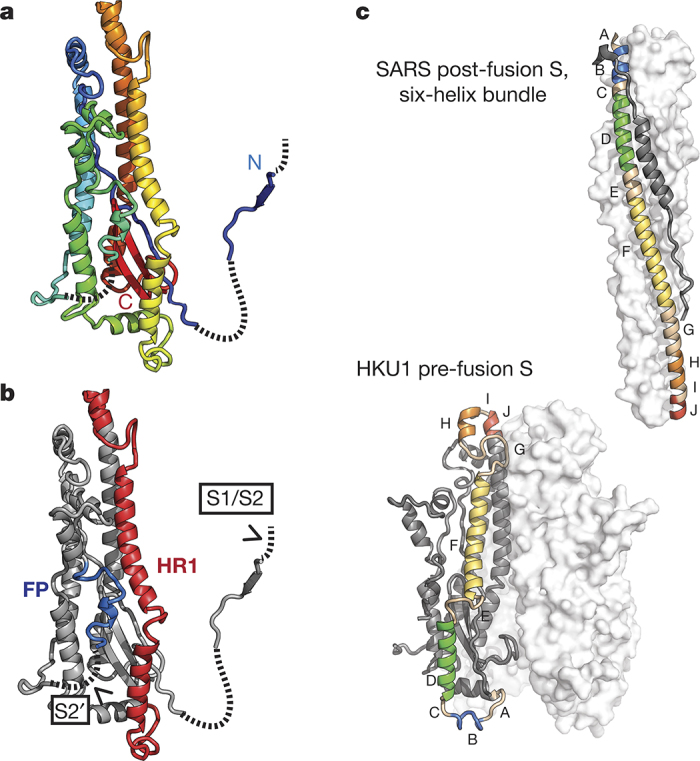
a, The HKU1 S2 subunit is coloured like a rainbow from the N-terminal β-strand (blue), which participates in S1 sub-domain 2, to the C terminus (red) before HR2. b, The HKU1 S2 structure contains the fusion peptide (FP) and a heptad repeat (HR1). Protease-recognition sites are indicated within disordered regions of the protein (dashed lines). c, A comparison of coronavirus S2 HR1 in the pre- and post-fusion22 conformations. Five HR1 α-helices are labelled and coloured like a rainbow from blue to red, N to C terminus, respectively. The structures are oriented to position similar portions of the central helix (red).
As in all class I viral fusion proteins, the coronavirus S2 subunit contains the four elements required for membrane fusion: a fusion peptide or loop, two heptad repeats (HR1 and HR2), and a transmembrane domain14,20,21. Refolding of HR1 into a long α-helix thrusts the fusion peptide into the host-cell membrane, and as the two heptad repeats interact to form a coiled-coil, the host and viral membranes are brought together. The fusion peptide, conserved among coronavirus S proteins17 (Extended Data Fig. 6), is located on the exterior of the HKU1 S protein and is adjacent to the putative S2′ cleavage site, which remains uncleaved in our structure. The fusion peptide forms a short helix and a loop, with most of the hydrophobic amino acids buried in an interface with other elements of S2. Unlike influenza HA where the C terminus of the fusion peptide is only 14 amino acids away from the N terminus of HR1, the fusion peptide of HKU1 S is 60 amino acids away from HR1. This span of protein contains four short α-helices and several longer regions lacking regular secondary structure. This intervening sequence is also buried beneath SD-2 and the S2′ cleavage site, suggesting that cleavage may affect the proclivity of S2 for undergoing the transition to the post-fusion conformation.
Coronavirus S protein heptad repeats are unusually large with HR1 encompassing more than 90 amino acids20. In the cryo-EM structure, HR2 is located at the base of the HKU1 S protein near the viral membrane, but is poorly ordered, precluding unambiguous assignment of the residues. However, HR1 is well ordered and arranged along the length of the S2 subunit, forming four short helices and part of the central three-helix bundle. This arrangement of HR1 is similar to that of influenza HA, although in HA the HR1 is organized as two helices connected by a long loop14. Conversion of influenza HA to the post-fusion conformation requires these protein elements to transition into a single long α-helix21. The post-fusion six-helix bundle structures of SARS-CoV and MERS-CoV S2 heptad repeats22,23 reveal that coronavirus S proteins also undergo a similar transition (Fig. 3c). However, the S protein must carry out five such loop-to-helix transitions, highlighting the complexity of S proteins relative to other class I fusion proteins. In addition, the membrane distal regions of the pre-fusion S2 central three-helix bundle (S2 amino acids 1070–1076), which is the C-terminal portion of HR1, are splayed outwards from the threefold axis (Extended Data Fig. 7). In the available coronavirus post-fusion HR1–HR2 structures, this portion of HR1 forms a tight three-helix bundle22,23. Formation of this three-helix bundle may be prevented by interactions between the C-terminal end of the S2 HR1 and the S1 CTD, and thus disruption of these interactions through receptor-induced conformational changes would provide an additional means by which receptor binding in S1 can initiate S2-mediated membrane fusion. Indeed, protease cleavage and an acidic pH are thought to be insufficient to trigger the transition to the post-fusion conformation without additional destabilization provided by receptor binding24,25,26.
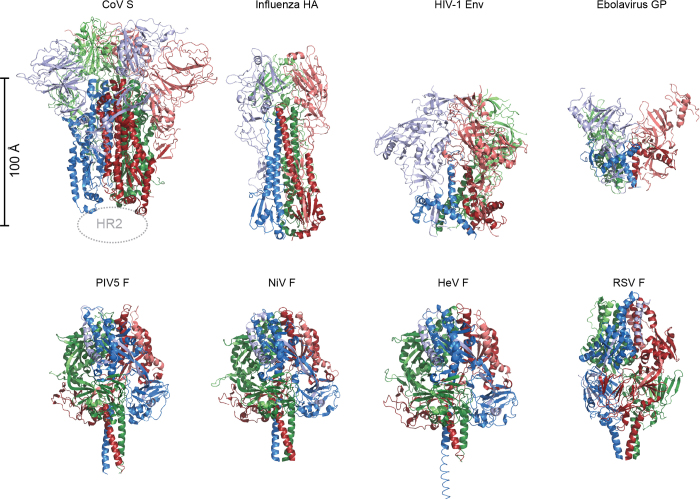
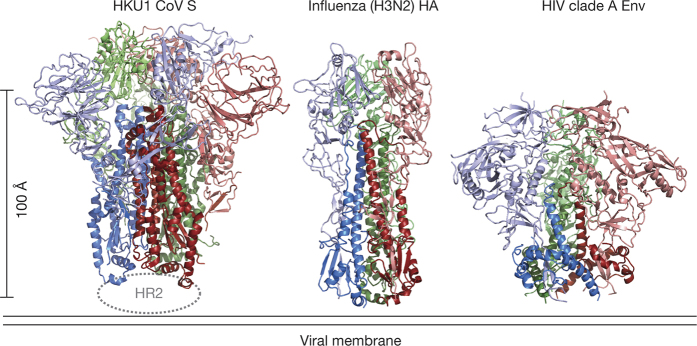
The formation of anti-parallel six-helix bundles composed of HR1 and HR2 in the post-fusion conformation is a unifying feature of class I viral fusion proteins. However, the pre-fusion conformations of this protein family are incredibly diverse in size and topology (Extended Data Fig. 8). The HKU1 S protein structure presented here most closely resembles influenza virus HA and HIV-1 Env (Fig. 4), which also have receptor-binding subunits that cap the central helix of the fusion subunit14,15,27,28. However, some core elements of the fusion machinery are conserved amongst all class I fusion proteins, including paramyxovirus F proteins.
Extended Data Figure 8. Class I viral fusion proteins.
All class I fusion proteins require proteolytic cleavage adjacent to the fusion peptide or loop, and the metastable pre-fusion state is triggered by a series of events that involve pH change or receptor binding. The post-fusion conformations all contain anti-parallel six-helix bundles composed of the HR1 and HR2 from the membrane-proximal subunit. However, there is a great diversity in pre-fusion conformations as shown here. Members of this class that also participate in receptor binding14,15,16,28,53 (top row), including S glycoproteins of coronaviruses, are organized such that their receptor binding subunits sit atop the fusion machinery, and need to be shed in order for membrane fusion to proceed. Paramyxovirus F proteins54,55,56,57 (bottom row) have a different architecture than the capped fusion proteins on the top row. The F proteins all have disulfide bonds between the membrane proximal and membrane distal subunits, and the two subunits remain interconnected throughout the rearrangement process.
Figure 4. Comparison of structurally related class I viral fusion proteins.
The fusion proteins from coronaviruses, influenza virus and HIV-1 are cleaved into receptor-binding subunits (pink, light green, light blue) and the viral fusion machinery (dark red, dark green, blue)14,15,16,28. Comparison to other class I fusion proteins can be found in Extended Data Fig. 8.
The HCoV-HKU1 S protein trimer in a pre-fusion conformation is, to our knowledge, the largest class I viral fusion glycoprotein structure determined to date (Fig. 4 and Extended Data Figs 8 and 9). Since betacoronavirus S proteins are similar in size and have a conserved domain organization, our findings should be generally applicable to other betacoronaviruses, including SARS-CoV and MERS-CoV (Extended Data Fig. 6). Our studies provide a structural basis for S protein function wherein the pre-fusion S protein is progressively matured and destabilized by receptor binding and protease cleavage. Following dissociation of the S1 subunits, HR1 would transition to a long α-helix, and the fusion peptide would be released from the side of the S2 subunit and inserted into host membranes. The structure and mechanistic insights presented here should enable engineering of pre-fusion stabilized coronavirus S proteins as vaccine immunogens against current and emerging betacoronaviruses, similar to recent efforts for other viral fusion proteins29,30. This work also acts as a springboard for future studies to define mechanisms of antibody recognition and neutralization, which will lead to an improved understanding of coronavirus immunity.
Extended Data Figure 9. HKU1 S glycosylation.
a, Sites of N-linked glycosylation on the HKU1 S trimer and b, a single monomer. Of the 30 potential N-linked glycosylation sites in a single protomer, the asparagine residues are observed for 21 sites and of these a small portion of density in the EM map is observed for 10 sites corresponding to the EndoH-trimmed sugars. Asparagines where glycan density is observed are shown as magenta spheres. Asparagines lacking glycan density are shown in green.
Methods
Data reporting
No statistical methods were used to predetermine sample size. The investigators were not blinded to allocation during experiments and outcome assessment.
Protein expression and purification
A mammalian-codon-optimized gene encoding HKU1 S (isolate N5, NCBI accession Q0ZME7) residues 1–1276 with a C-terminal T4 fibritin trimerization domain, a HRV3C cleavage site, and a 6xHis-tag was synthesized and subcloned into the eukaryotic expression vector pVRC8400. The S1/S2 furin-recognition site 752-RRKRR-756 was mutated to GGSGS to generate the uncleaved construct used for cryoEM studies. Three hours after this plasmid was transfected into FreeStyle 293-F cells (Invitrogen), kifunensine was added to a final concentration of 5 μM. FreeStyle 293-F cells are a high-transfection-efficiency cell line adapted for suspension culture derived from low passage clonal cultures and after purchase were not further authenticated. Cells were not confirmed to be free of mycoplasma, but were only used for protein expression. Cultures were harvested after six days, and protein was purified from the medium using Ni-NTA Superflow resin (Qiagen). The buffer was then exchanged using a HiPrep 26/10 desalting column (GE Healthcare Biosciences) from a high-imidazole elution buffer to a low pH buffer (20 mM Bis-Tris pH 6.5, 150 mM NaCl). Afterward, endoglycosidase H (EndoH) (10% w/w) and HRV3C protease (1% w/w) were added to the protein and the reaction was incubated overnight at 4 °C. The digested protein was further purified using a Superose 6 16/70 column (GE Healthcare Biosciences).
The furin-cleaved HKU1 S construct analysed by negative-stain EM was similar to the one described above except that it encoded residues 1–1249 and contained the wild-type RRKRR furin-recognition site. Expression and purification were also similar, except that a plasmid expressing furin was co-transfected into the FreeStyle 293-F cells to ensure complete processing of the protein.
Sample preparation for negative-stain electron microscopy
HKU1 S proteins were placed directly onto 400 copper mesh grids and then stained with 1% uranyl formate. Tris-buffered saline (TBS) was used as buffer if dilution was necessary.
Negative-stain electron microscopy data collection
Grids were loaded into a Tecnai T12 Spirit operating at 120 keV and imaged using a Tietz TemCam-F416 CMOS at 52,000 × magnification at ~1.5 μm under focus. Micrographs were collected using Leginon31 and processed within Appion32. Particles were picked using a difference-of-Gaussians approach33 and aligned using reference-free 2D classification employing iterative multivariate statistical analysis/multi-reference alignment (MRA/MSA) using a binning factor of 2 to remove amorphous particles34. Particles in classes that did not represent views of HKU1 S proteins were discarded. ISAC35 was used to generate a template stack from which initial 3D models were generated using the EMAN2 (ref. 36) procedure initialmodel.py. 3D models were refined using EMAN1 (ref. 37).
Sample preparation for cryo-electron microscopy
Sample solution (3 μl) was applied to the carbon face of a CF-2/2-4C C-Flat grid (Electron Microscopy Sciences, Protochips) that had been plasma cleaned for five seconds using a mixture of Ar/O2 (Gatan Solarus 950 Plasma system). The grid was then manually blotted and immediately plunged into liquid ethane using a manual freeze plunger.
Cryo-electron microscopy data collection
Movies were collected via the Leginon interface on a FEI Titan Krios operating at 300 keV mounted with a Gatan K2 direct-electron detector31. Each movie was collected in counting mode at 22,500 × nominal magnification resulting in a calibrated pixel size of 1.31 Å/pix at the object level. A dose rate of ~10 e−/((cam pix) × s) was used; exposure time was 200 ms per frame. The data collection resulted in a total of 1,049 movies containing 50 frames each. Total dose per movie was 57 e−/Å2. Data were collected at 1.0 to 3.5 μm under focus.
Cryo-electron microscopy data processing
Frames in each movie were aligned38, and CTF estimation was carried out using CTFFIND3 (ref. 39). Particles were picked from a subset of the data employing a difference-of-Gaussians approach33 and aligned using reference-free 2D classification employing iterative MRA/MSA using a binning factor of two34. The resulting 2,188 particles were used to generate an initial 25 Å lowpass-filtered 3D reconstruction using EMAN2. SPIDER refproj.spi40 with a delta theta angle of 15 degrees was used to generate 83 projection images of the initial 3D reconstruction. These projection images were used as templates for picking particles from the entire cryo data set. Particles from the entire data set were aligned and classified with the same methods used for the subset of particles stated above. After 2D classification, unbinned selected particles were symmetrically refined in RELION version 1.3 (refs 41, 42) against the initial 3D reconstruction filtered to 60 Å resolution. This refinement was followed by particle polishing and refinement of the resulting realigned, B-factor-weighted and signal-integrated particles using RELION version 1.4b1. The resolution of the final map was 4.04 Å at an FSC cutoff of 0.143. A mask was generated in RELION using a threshold that accounted for the entire structure. From this threshold, the mask was further dilated by 3 voxels and a Gaussian fall-off was generated over an additional 6 voxels. The mask effect on FSC was taken into consideration. Phases were randomized in the unfiltered half-set maps for initial FSC lower than 0.8 and a new FSC between these phase-randomized maps was generated and used to correct for mask effects in the final FSC-based resolution estimate. The reported resolution of 4.04 Å is the RELION CorrelationCorrected value
The map was B-factor sharpened employing FSC-weighting. The B-factor was estimated in RELION based on the resolution range from 10 Å to 2.62 Å (B-factor = −117 Å2). The detector MTF file was provided to RELION.
Model building and refinement
An initial model of the S1 NTD was generated using the Modeller43 homology modelling tool in UCSF Chimera44 with the BCoV NTD (PDB 4H14)8 as a template. The NTD homology model was docked into the HKU1 S protein EM density and refined with Rosetta density-guided iterative local refinement45 while imposing C3 symmetry. Rosetta output models were clustered based on pairwise r.m.s.d. using a cluster radius of 2.15 Å. The lowest energy model from the largest cluster was selected for additional refinement. This model and the conserved CTD core from SARS-CoV (PDB 2AJF)11 were used as starting structures for model building and refinement. These starting models and the remaining HKU1 protein sequence were modelled manually using COOT46 and refined using RosettaRelax47. Structures were evaluated using EMRinger48 and Molprobity49. Figures were produced in the PyMol50 or UCSF Chimera44 software packages.
Acknowledgements
We thank colleagues and members of our laboratories for critical reading of the manuscript. This work was supported by start-up funds to A.B.W. from the Scripps Research Institute and to J.S.M from the Geisel School of Medicine at Dartmouth, and funding from intramural NIAID to B.S.G. Partial salary support for R.N.K. was provided by NIH grant R56 AI118016. This is manuscript 29286 from TSRI.
Extended data figures and tables
PowerPoint slides
Author Contributions
H.M.Y. and K.S.C. designed the HKU1 S expression constructs. N.W. expressed and purified the proteins. C.A.C., H.L.T., and J.P. prepared samples for cryo-EM, collected images, and processed the data. R.N.K. and C.A.C built and refined the model. B.S.G., J.S.M., and A.B.W. conceived of the project. R.N.K, C.A.C., N.W., B.S.G, J.S.M and A.B.W analysed the results and wrote the manuscript, with all authors editing and approving the final manuscript.
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
The negative stain EM maps of the HKU1 spike ectodomain have been deposited in the EMDB under accession numbers EMD-8066, EMD-8067, and EMD-8068. The cryo-EM map of the HKU1 spike ectodomain has been deposited in the EMDB under accession number EMD-8069 and the coordinates of the HKU1 spike ectodomain structure have been deposited in the PDB under accession code 5I08.
Competing interests
The authors declare no competing financial interests.
Footnotes
Robert N. Kirchdoerfer and Christopher A. Cottrell: These authors contributed equally to this work.
Contributor Information
Jason S. McLellan, Email: Jason.S.McLellan@Dartmouth.edu
Andrew B. Ward, Email: ABWard@Scripps.edu
References
- 1.Woo PC, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christian MD, Poutanen SM, Loutfy MR, Muller MP, Low DE. Severe acute respiratory syndrome. Clin. Infect. Dis. 2004;38:1420–1427. doi: 10.1086/420743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 4.Graham RL, Baric RS. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li W, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mou H, et al. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J. Virol. 2013;87:9379–9383. doi: 10.1128/JVI.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng G, et al. Crystal structure of bovine coronavirus spike protein lectin domain. J. Biol. Chem. 2012;287:41931–41938. doi: 10.1074/jbc.M112.418210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang X, et al. Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J. Virol. 2015;89:7202–7213. doi: 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 2015;89:1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 12.Lu G, et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian Z, et al. Identification of the receptor-binding domain of the spike glycoprotein of human betacoronavirus HKU1. J. Virol. 2015;89:8816–8827. doi: 10.1128/JVI.03737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 15.Julien JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyumkis D, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madu IG, Roth SL, Belouzard S, Whittaker GR. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 2009;83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schornberg K, et al. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmer G, Budz L, Herrler G. Proteolytic activation of respiratory syncytial virus fusion protein. Cleavage at two furin consensus sequences. J. Biol. Chem. 2001;276:31642–31650. doi: 10.1074/jbc.M102633200. [DOI] [PubMed] [Google Scholar]
- 20.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 22.Duquerroy S, Vigouroux A, Rottier PJ, Rey FA, Bosch BJ. Central ions and lateral asparagine/glutamine zippers stabilize the post-fusion hairpin conformation of the SARS coronavirus spike glycoprotein. Virology. 2005;335:276–285. doi: 10.1016/j.virol.2005.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu L, et al. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nature Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li F, et al. Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J. Virol. 2006;80:6794–6800. doi: 10.1128/JVI.02744-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuyama S, Taguchi F. Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis. J. Virol. 2009;83:11133–11141. doi: 10.1128/JVI.00959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simmons G, et al. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl Acad. Sci. USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong L, et al. Complete epitopes for vaccine design derived from a crystal structure of the broadly neutralizing antibodies PGT128 and 8ANC195 in complex with an HIV-1 Env trimer. Acta Crystallogr. D. 2015;71:2099–2108. doi: 10.1107/S1399004715013917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee PS, et al. Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nature Commun. 2014;5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLellan JS, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanders RW, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Lander GC, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogura T, Iwasaki K, Sato C. Topology representing network enables highly accurate classification of protein images taken by cryo electron-microscope without masking. J. Struct. Biol. 2003;143:185–200. doi: 10.1016/j.jsb.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20:237–247. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 38.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 40.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 41.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nature Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics. 2014;47:5.6.1–5.6.32. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- 44.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.DiMaio F, et al. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nature Methods. 2015;12:361–365. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DiMaio F, et al. Refinement of protein structures into low-resolution density maps using Rosetta. J. Mol. Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barad BA, et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nature Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schrodinger, L. The PyMOL Molecular Graphics System, Version 1.5.0.4. (2012)
- 51.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nature Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sievers F, Higgins DG. Clustal Omega. Curr Protoc Bioinformatics. 2014;48:3.13.11–13.13.16. doi: 10.1002/0471250953.bi0313s48. [DOI] [PubMed] [Google Scholar]
- 53.Lee JE, et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLellan JS, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welch BD, et al. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc. Natl Acad. Sci. USA. 2012;109:16672–16677. doi: 10.1073/pnas.1213802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong JJ, Paterson RG, Lamb RA, Jardetzky TS. Structure and stabilization of the Hendra virus F glycoprotein in its prefusion form. Proc. Natl Acad. Sci. USA. 2016;113:1056–1061. doi: 10.1073/pnas.1523303113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu K, et al. Crystal structure of the pre-fusion Nipah virus fusion glycoprotein reveals a novel hexamer-of-trimers assembly. PLoS Pathog. 2015;11:e1005322. doi: 10.1371/journal.ppat.1005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
The negative stain EM maps of the HKU1 spike ectodomain have been deposited in the EMDB under accession numbers EMD-8066, EMD-8067, and EMD-8068. The cryo-EM map of the HKU1 spike ectodomain has been deposited in the EMDB under accession number EMD-8069 and the coordinates of the HKU1 spike ectodomain structure have been deposited in the PDB under accession code 5I08.